Non-coding cause of congenital heart defects
Non-coding cause of congenital heart defects: Abnormal RNA splicing with multiple isoforms as a mechanism for heterotaxy
HGG Adv. 2024 Oct 10;5(4):100353. doi: 10.1016/j.xhgg.2024.100353. Epub 2024 Sep 12.
John R Wells, Maria B Padua, Allison M Haaning, Amanda M Smith, Shaine A Morris, Muhammad Tariq, Stephanie M Ware.
Click here to view article at HGG Advances.
Click here to view article at Pubmed.
Click here to view article on Xenbase.
Abstract
Heterotaxy is a disorder characterized by severe congenital heart defects (CHDs) and abnormal left-right patterning in other thoracic or abdominal organs. Clinical and research-based genetic testing has previously focused on evaluation of coding variants to identify causes of CHDs, leaving non-coding causes of CHDs largely unknown. Variants in the transcription factor zinc finger of the cerebellum 3 (ZIC3) cause X-linked heterotaxy. We identified an X-linked heterotaxy pedigree without a coding variant in ZIC3. Whole-genome sequencing revealed a deep intronic variant (ZIC3 c.1224+3286A>G) predicted to alter RNA splicing. An in vitro minigene splicing assay confirmed the variant acts as a cryptic splice acceptor. CRISPR-Cas9 served to introduce the ZIC3 c.1224+3286A>G variant into human embryonic stem cells demonstrating pseudoexon inclusion caused by the variant. Surprisingly, Sanger sequencing of the resulting ZIC3 c.1224+3286A>G amplicons revealed several isoforms, many of which bypass the normal coding sequence of the third exon of ZIC3, causing a disruption of a DNA-binding domain and a nuclear localization signal. Short- and long-read mRNA sequencing confirmed these initial results and identified additional splicing patterns. Assessment of four isoforms determined abnormal functions in vitro and in vivo while treatment with a splice-blocking morpholino partially rescued ZIC3. These results demonstrate that pseudoexon inclusion in ZIC3 can cause heterotaxy and provide functional validation of non-coding disease causation. Our results suggest the importance of non-coding variants in heterotaxy and the need for improved methods to identify and classify non-coding variation that may contribute to CHDs.
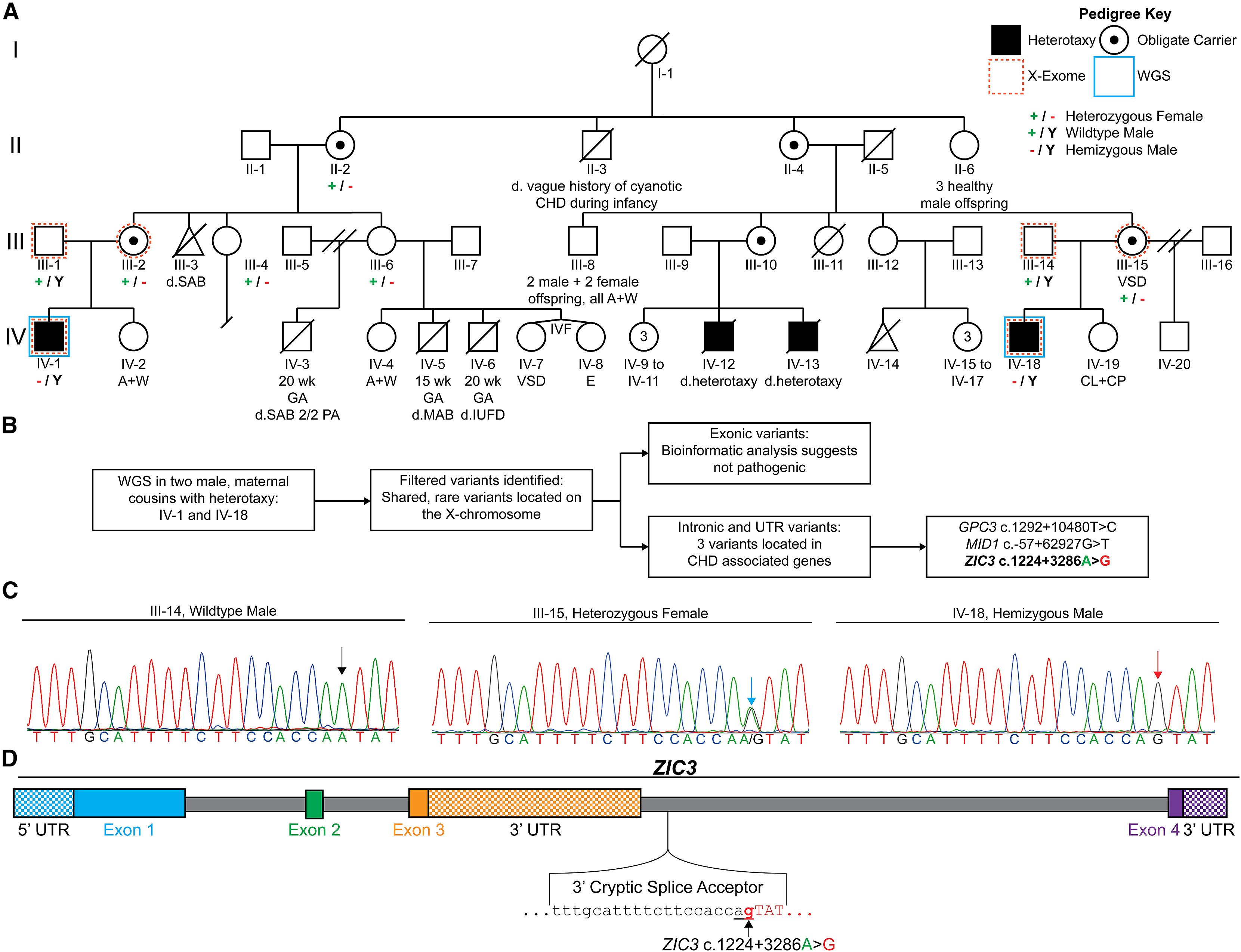
Figure 1. Intronic variant in ZIC3 identified by WGS of an X-linked heterotaxy pedigree.
(A) A pedigree with four heterotaxy-affected males (black squares) displaying an X-linked recessive inheritance pattern. Generations are labeled I–IV while individuals within each generation are numbered from 1 to 20. Deceased individuals are denoted with a diagonal line with the cause of death indicated as “d.” when available. X-exome sequencing (dashed orange outer frame) performed on two separate trios did not identify a coding variant in ZIC3, while WGS (solid blue outer frame) completed on two males with heterotaxy (IV-1 and IV-18) revealed a ZIC3 c.1224+3286A>G intronic variant. A + W, alive and well; CHD, congenital heart defect; CL, cleft lip; CP, cleft palate; E, encephalocele; GA, gestational age; IUFD, intrauterine fetal demise; IVF, in vitro fertilization; MAB, missed abortion; P, pregnancy; SAB, spontaneous abortion; SAB 2/2 PA, spontaneous abortion secondary to placental abruption; VSD; ventricular septal defect; wk, week.
(B) Schematic diagram of IV-1 and IV-18 WGS and variant filtering steps (Figure S1) identifying ZIC3 c.1224+3286A>G as a plausible disease-causing variant.
(C) Sanger sequencing chromatogram of the predicted 3′ splice acceptor site in III-14, III-15, and IV-18. The sequence at the ZIC3 c.1224+3286 position is denoted as a black arrow for the father (ZIC3 c.1224+3286A), a red arrow for the hemizygous male with heterotaxy (ZIC3 c.1224+3286A>G), and a blue arrow for the heterozygous mother.
(D) ZIC3 contains four exons with the untranslated regions (UTRs) shown as checkerboard-colored rectangles. The ZIC3 c.1224+3286A>G variant is located within the intronic region between exons 3 and 4 and it is predicted to result in a cryptic 3′ splice acceptor sequence (see also Table S6). Predicted intronic and exonic sequences are shown in lowercase and capital letters, respectively. The mutated “g” in ZIC3 c.1224+3286A>G is shown in bold red while the predicted “ag” cryptic splice acceptor caused by the variant is underlined.
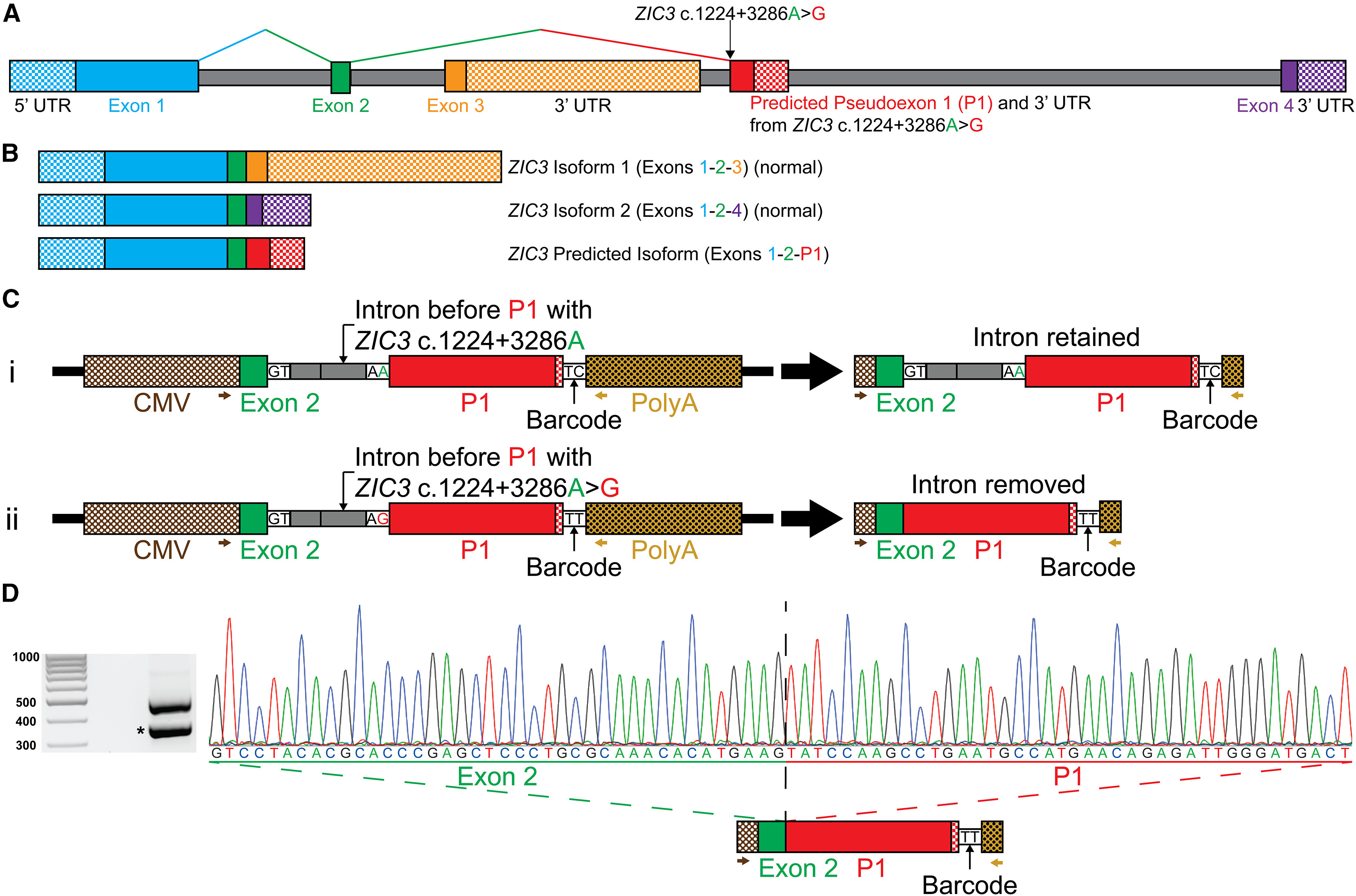
Figure 2. The ZIC3 c.1224+3286A>G variant acts as a 3′ splice acceptor between exon 2 and the predicted P1 in a minigene assay.
(A) Illustrative representation of ZIC3 showing the ZIC3 c.1224+3286A>G variant predicted to result in a 3′ cryptic splice acceptor that causes the inclusion of P1.
(B) ZIC3 encodes two isoforms: isoform 1 formed by exons 1-2-3 (dominant isoform), and isoform 2 encoded by exons 1-2-4. The ZIC3 predicted isoform is expected to be encoded by exons 1-2-P1.
(C) Minigene plasmids contain a 1,289-bp minigene construct composed of a CMV promoter (brown and white checkerboard pattern), exon 2 (final 40-bp portion, green), intron (140 bp, gray), P1 (255 bp, predicted coding region of P1, red; predicted 3′ UTR region of P1, red and white checkerboard pattern), a 2-bp barcode sequence, and an SV40 poly(A) tail signal sequence (dark yellow and white checkerboard pattern). (Ci) The exon 2 to P1 control construct contains the reference ZIC3 c.1224+3286A sequence predicted to not alter splicing resulting in full intron retention. (Cii) The exon 2 to P1 construct containing the ZIC3 c.1224+3286A>G variant is predicted to generate a 3′ splice acceptor and remove the intron.
(D) Electrophoretogram of amplicons obtained by RT-PCR from amplified cDNA of the ZIC3 c.1224+3286A>G variant construct, using primers located on the CMV promoter (brown arrow) and the SV40 poly(A) tail sequence (dark yellow arrow). The top amplicon (∼500 bp) corresponds to full intron retention, while the ∼360-bp amplicon represents intron removal. Sanger sequencing chromatogram of the ∼360-bp amplicon showing that the intronic region between exon 2 and P1 was removed.
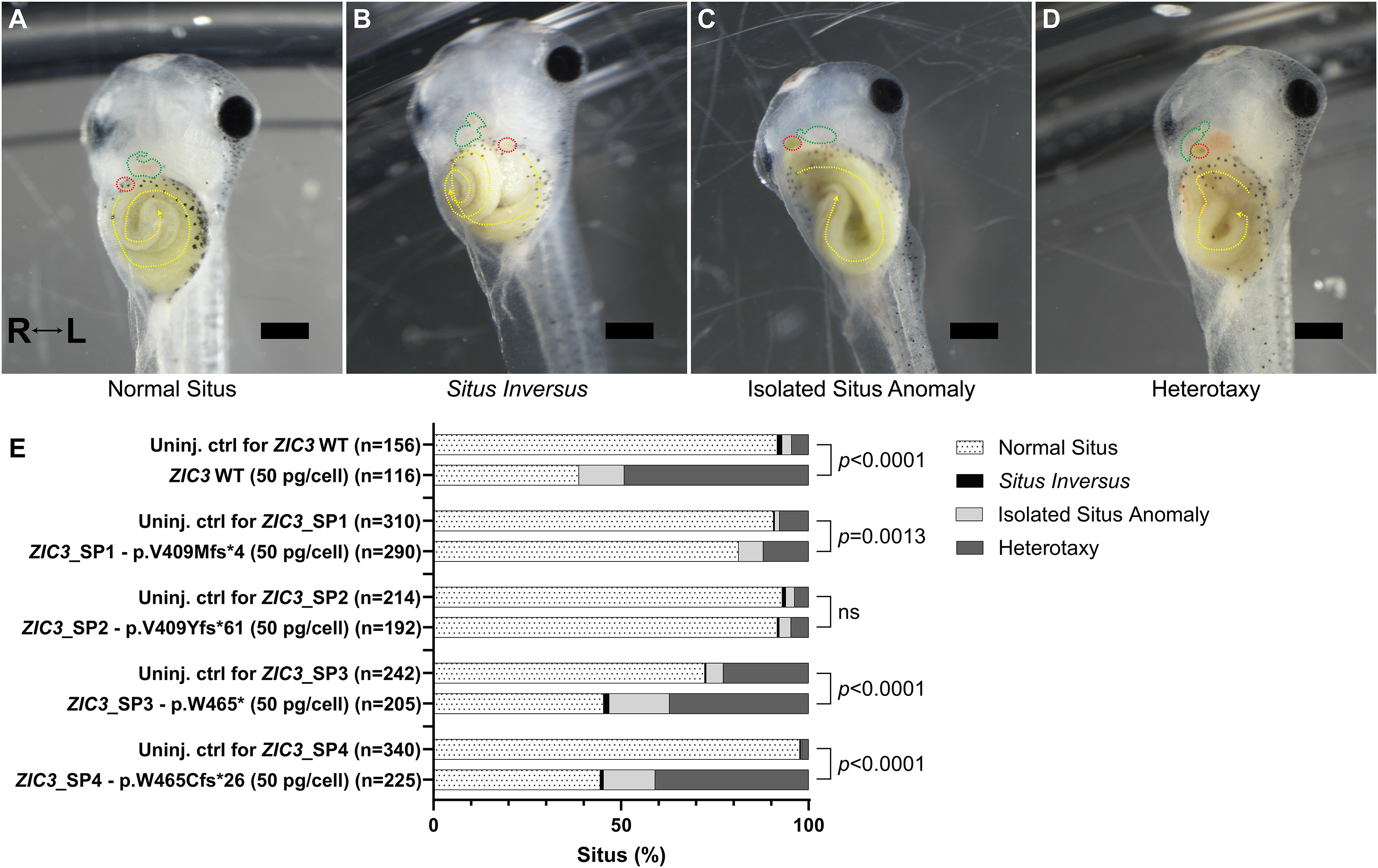
Figure 8. Situs abnormalities in X. laevis embryos injected with abnormal ZIC3 isoforms.
(A–D) Representative images of X. laevis tadpoles (stage 47) that received at the two-cell stage one of the following: no injection (uninjected control [uninj. ctrl]) (E) in vitro synthesized mRNA encoding the coding sequence of HA-tagged ZIC3 isoform 1 (ZIC3 WT, 50 pg/cell; 100 pg/embryo), in vitro synthesized mRNA encoding the coding sequence of one of the four HA-tagged ZIC3 c.1224+3286A>G isoforms (50 pg/cell; 100 pg/embryo). Situs defects were assessed by the position of the heart, gallbladder, and gut and categorized into one of four groups. (A) Normal situs tadpoles display normal heart looping (green dashed line), normal right gut origin and counterclockwise gut coil (yellow dashed line), and normal position of the gallbladder on the right (red dashed line). (B) Situs inversus tadpoles exhibit reversed heart looping, left gut origin with clockwise gut coil, and leftward gallbladder. (C) Isolated situs anomaly tadpoles have one organ defect (right-origin gut coil with clockwise rotation), while (D) heterotaxy tadpoles have two or more organ defects (reversed heart looping, a left gallbladder position, and a left gut origin with counterclockwise gut coil). Scale bars, 0.5 mm. Videos are provided as Videos S5, S6, S7, and S8. The Fisher’s exact test (two sided) served to calculate significance (p < 0.05) by comparing the number of embryos with normal situs to the sum of the number of embryos with abnormal situs (situs inversus, isolated situs anomaly, and heterotaxy). Raw counts used for statistical analysis are included in Table S7. ns, not significant.
Adapted with permission from Cell Press on behalf of HGG Advances: Wells et al. (2024). Non-coding cause of congenital heart defects: Abnormal RNA splicing with multiple isoforms as a mechanism for heterotaxy. HGG Adv. 2024 Oct 10;5(4):100353. doi: 10.1016/j.xhgg.2024.100353. Epub 2024 Sep 12.
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
Last Updated: 2024-10-16