Cerebellar granular neuron progenitors exit their germinative niche via BarH-like1 activity mediated partly by inhibition of T-cell factor
Development 2024 Jul 01;15113:. doi: 10.1242/dev.202234.
Bou-Rouphael J, Doulazmi M, Eschstruth A, Abdou A, Durand BC.
Click here to view article at Development.
Click here to view article at Pubmed.
Click here to view article on Xenbase.
Abstract
Cerebellar granule neuron progenitors (GNPs) originate from the upper rhombic lip (URL), a germinative niche in which developmental defects produce human diseases. T-cell factor (TCF) responsiveness and Notch dependence are hallmarks of self-renewal in neural stem cells. TCF activity, together with transcripts encoding proneural gene repressors hairy and enhancer of split (Hes/Hey), are detected in the URL; however, their functions and regulatory modes are undeciphered. Here, we established amphibian as a pertinent model for studying vertebrate URL development. The amphibian long-lived URL is TCF active, whereas the external granular layer (EGL) is non-proliferative and expresses hes4 and hes5 genes. Using functional and transcriptomic approaches, we show that TCF activity is necessary for URL emergence and maintenance. We establish that the transcription factor Barhl1 controls GNP exit from the URL, acting partly through direct TCF inhibition. Identification of Barhl1 target genes suggests that, besides TCF, Barhl1 inhibits transcription of hes5 genes independently of Notch signaling. Observations in amniotes suggest a conserved role for Barhl in maintenance of the URL and/or EGL via co-regulation of TCF, Hes and Hey genes.
The upper rhombic lip, an important cerebellar germinative niche, has recently been a subject of intense investigation as defects in the human URL developmental program leads to serious diseases. The upper rhombic lip generates cerebellar granule neuron, the predominant neuronal population in the entire Central Nervous System. T-Cell Factors are the main transcriptional mediators of what is known as the Wnt/β-catenin cell-to-cell signaling pathway, one of the most conserved in the animal kingdom. T-Cell Factors responsiveness is a hallmark of self-renewal in neural stem cells. Whereas T-Cell Factors activity is detected in the upper rhombic lip, its functions and regulatory modes were previously undeciphered.
Here we established amphibians as a new and pertinent model system to study vertebrate cerebellum and upper rhombic lip development, together with their potentially harmful developmental aberrations. Using functional and transcriptomic approaches, we show that T-Cell Factor activity is strictly necessary for upper rhombic lip emergence and maintenance. The transcription factor Barhl1 is the main gate keeper of granule neuron progenitors exit from their germinative niche acting through direct binding and silencing of T-Cell Factor transcriptional activity. Our identification of Barhl1 target genes shows that aside from T-Cell Factor activity, Barhl1 inhibits transcription of genes in the hairy enhancer of split (hes/hey) family, known to be required for neural stem cell maintenance. Our research has potential clinical implications since both T Cell Factors and a Barhl roles in emergence and maintenance of the upper rhombic lip appear conserved in mammals.
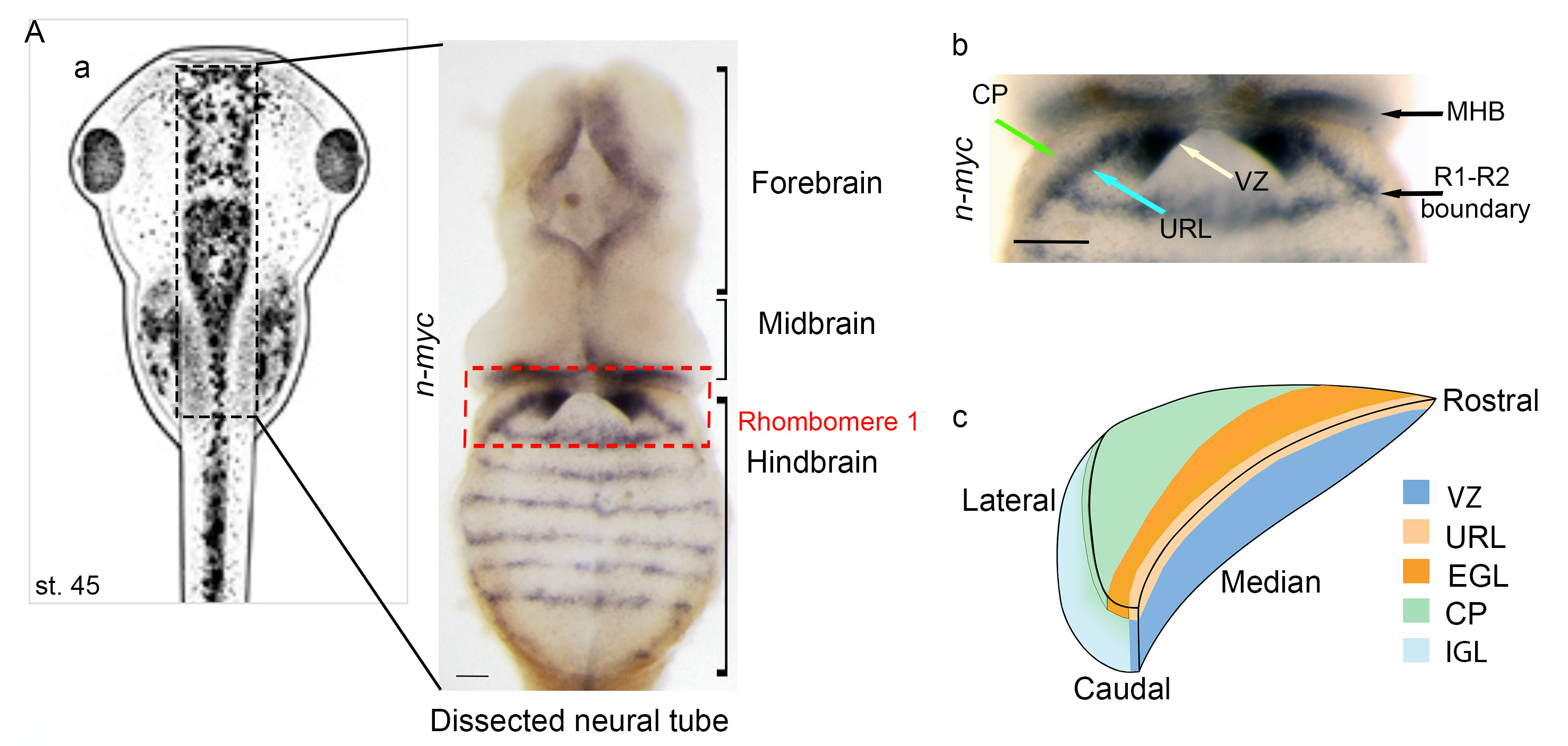
Figure 1A: Temporal and spatial expression pattern of genes involved in granule neuron progenitor development. (A) Neural tube dissection and analysis. (Aa) Representation of stage (st.) 45 X. laevis embryo. After in situ hybridization, neural tubes are dissected, as indicated on the right (entire neural tube) and in Ab, which focuses on rhombomere 1. The proliferation marker nmyc is expressed in the upper rhombic lip (URL) (blue arrow) and the ventricular zone VZ (white arrow). Red dotted line delineates rhombomere 1 (R1), which is located caudal to the midbrain-hindbrain boundary (MHB). nmyc marks proliferating progenitors at the boundary between R1 and R2, and is used as a marker of the caudal limit of the cerebellar primordium. The cerebellar plate (CP) is indicated (green arrow). (Ac) A stage 42 Xenopus half R1. (B-H) In situ hybridization analysis of granule neuron progenitor (GNP) markers in X. laevis embryos at the indicated Nieuwkoop and Faber stages. Dorsal (a,b), lateral (a′,b′).
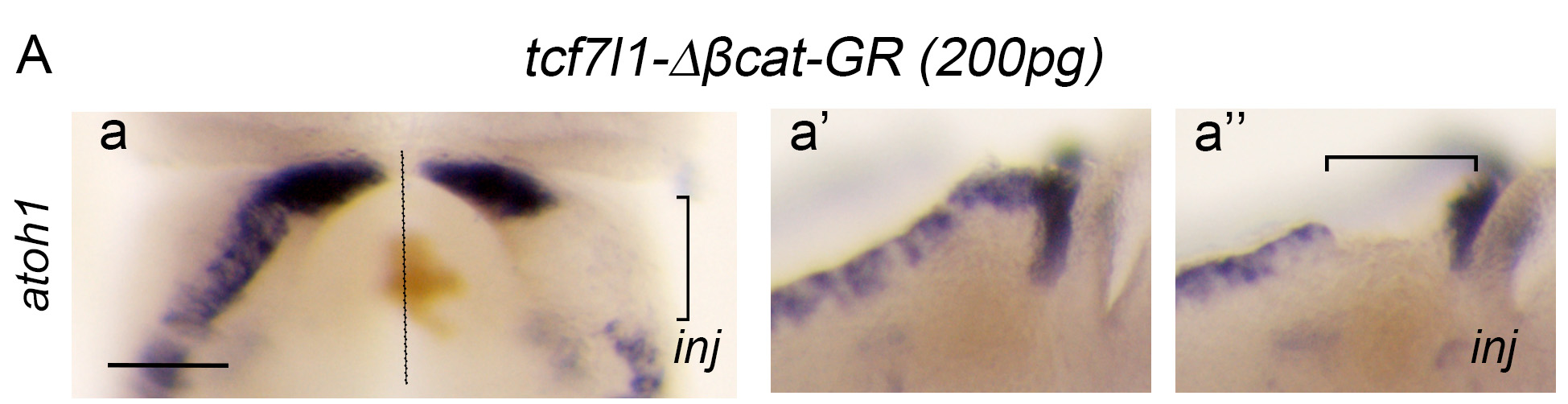
Figure 2A: TCF activity is required for the induction of the upper rhombic lip and its inhibition by Barhl1 is necessary for the proper progression of granule neuron progenitor development. (A) Overexpression of tcf7l1-Δβcat-GR inhibits and/or abolishes atoh1 expression in a dose-dependent manner. In situ hybridization analysis of atoh1 expression in rhombomere 1 (R1) showing dorsal views (a,b) and lateral views (a′,a′′,b′,b′′) of control sides (a′,b′) and injected sides (a′′,b′′) of stage 45 X. laevis embryos unilaterally injected with 200 pg (a-a′′) or 100 pg (b-b′′) of tcf7l1-Δβcat-GR. The non-injected side is an internal control.
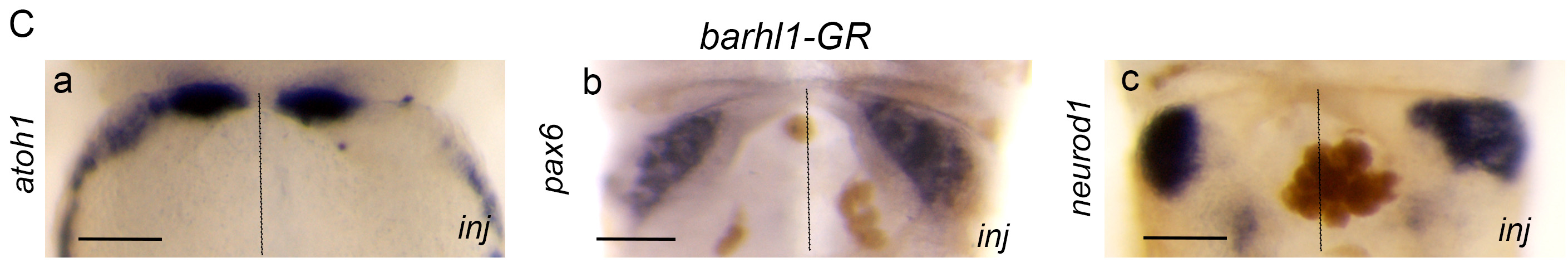
Figure 2C: TCF activity is required for the induction of the upper rhombic lip and its inhibition by Barhl1 is necessary for the proper progression of granule neuron progenitor development. (C) barhl1 overexpression phenocopies the defects of tcf7l1dn overexpression. Dorsal views showing atoh1, barhl1 and neurod1 (a-c) expression in R1 primordium of stage 45 X. laevis embryos injected with mBarhl1GR (200 pg).
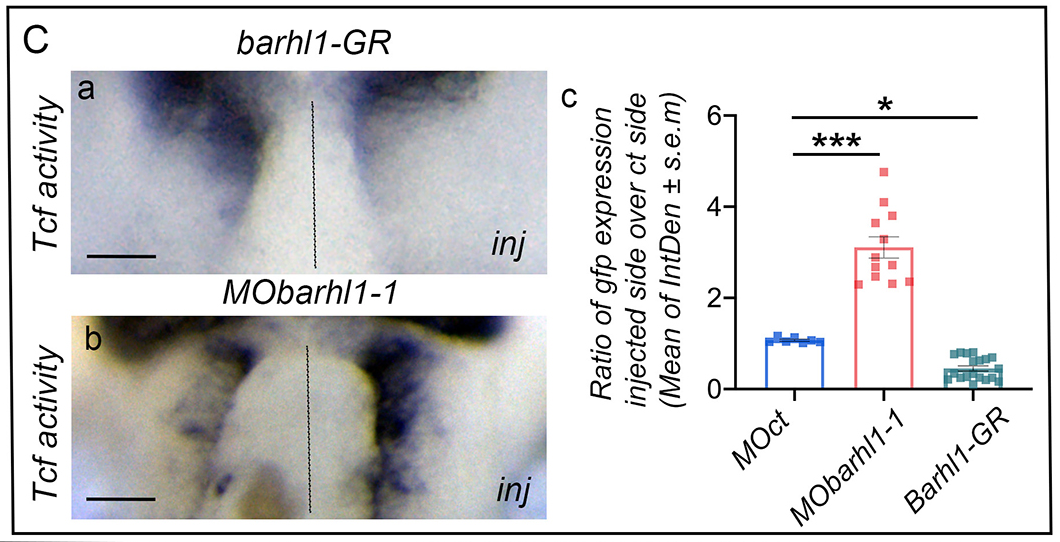
Figure 4C: Barhl1 physically interacts with Tcf7l1 and Gro, and limits TCF transcriptional activity. (C) Barhl1 limits TCF transcriptional activity in vivo. In situ hybridization analysis of gfp expression in X. tropicalis pbin7LefdGFP embryos injected either unilaterally with (Ca) mBarhl1GR (200 pg) and (Cb) MObarhl1-1, or before division with (Cc) Quantification. Integrated densities (IntDen) of marker expression are measured. The ratio of marker expression in injected side over control side is represented and indicated as mean±s.e.m. The result from each tadpole is represented by a square. Dotted lines separate injected and control sides. Scale bar: 150 μm. inj, injected side. Statistical analysis: (Cc) one-way ANOVA [F(2,35)=111,3; P<0.001] followed by post-hoc Tukey’s test: Data are presented as mean±s.e.m. *P≤0.05; ***P≤0.001.
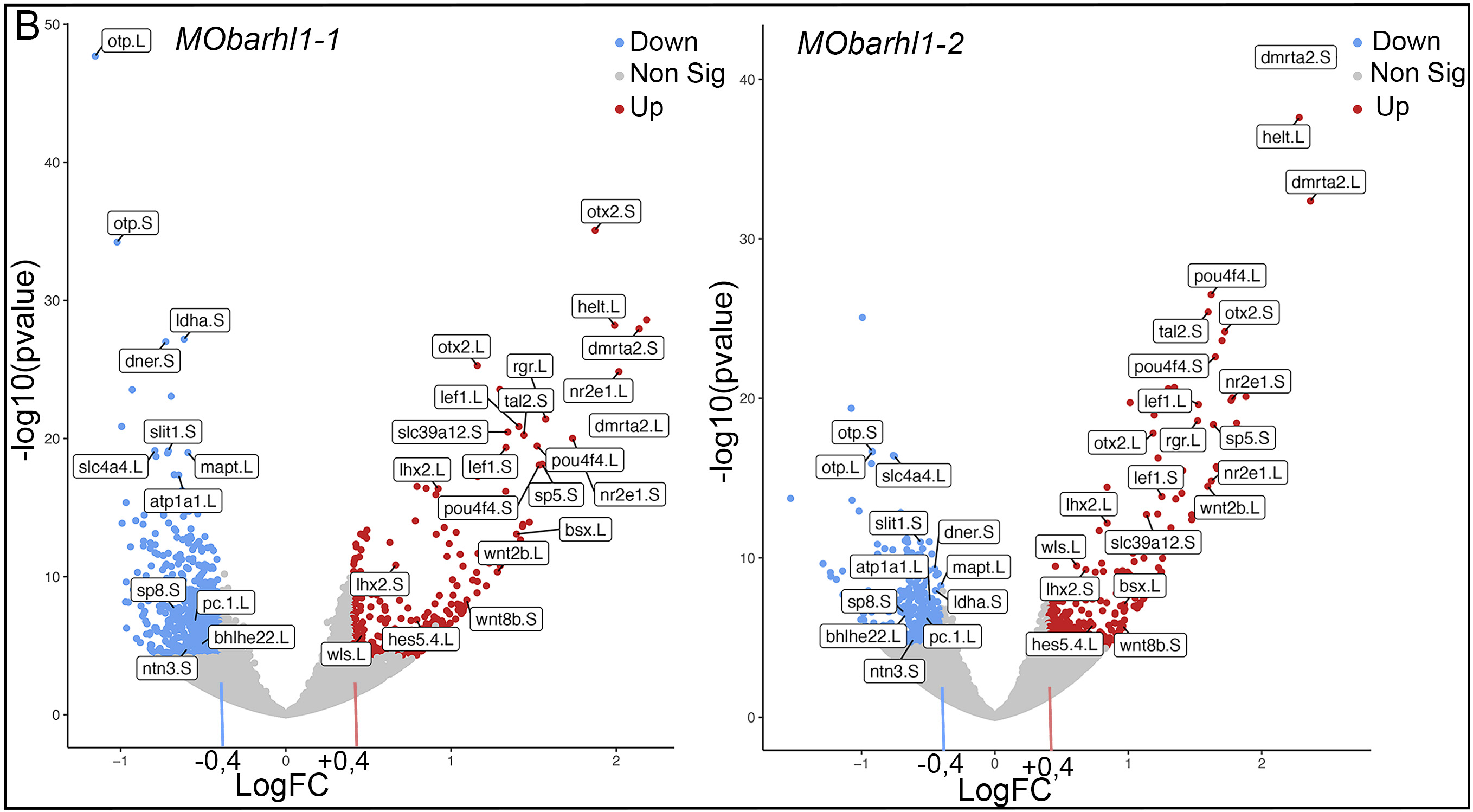
Figure 6B: RNA-sequencing data processing and analysis. (B) Volcano plots showing a selection of significant DEGs with pAdj<0.001 in (left) MObarhl1-1 versus MOct and (right) MObarhl1-2 versus MOct. Upregulated genes with Log2FC>0.4 and downregulated genes with Log2FC<−0.4 are shown. Red and blue dots indicate significant DEGs that are upregulated and downregulated, respectively. Grey dots indicate RNAs with non-significant differences. PCA and volcano plots were generated using Galaxy.
Image credits: Johnny Bou-Rouphael and Beatrice Durand.
Adapted with permission from The Company of Biologists on behalf of Development: Bou-Rouphael et al. (2024). Cerebellar granular neuron progenitors exit their germinative niche via BarH-like1 activity mediated partly by inhibition of T-cell factor. Development 2024 Jul 01;15113:. doi: 10.1242/dev.202234.
