The H2A.Z and NuRD associated protein HMG20A controls early head and heart developmental transcription programs
Nat Commun. 2023 Jan 28;14(1):472. doi: 10.1038/s41467-023-36114-x.
Andreas Herchenröther, Stefanie Gossen, Tobias Friedrich, Alexander Reim, Nadine Daus, Felix Diegmüller, Jörg Leers, Hakimeh Moghaddas Sani, Sarah Gerstner, Leah Schwarz, Inga Stellmacher, Laura Victoria Szymkowiak, Andrea Nist, Thorsten Stiewe, Tilman Borggrefe, Matthias Mann, Joel P. Mackay, Marek Bartkuhn, Annette Borchers, Jie Lan & Sandra B. Hake
Click here to view article at Nature Communications.
Click here to view article on PubMed.
Click here to view article on Xenbase.
Abstract
Specialized chromatin-binding proteins are required for DNA-based processes during development. We recently established PWWP2A as a direct histone variant H2A.Z interactor involved in mitosis and craniofacial development. Here, we identify the H2A.Z/PWWP2A-associated protein HMG20A as part of several chromatin-modifying complexes, including NuRD, and show that it localizes to distinct genomic regulatory regions. Hmg20a depletion causes severe head and heart developmental defects in Xenopus laevis. Our data indicate that craniofacial malformations are caused by defects in neural crest cell (NCC) migration and cartilage formation. These developmental failures are phenocopied in Hmg20a-depleted mESCs, which show inefficient differentiation into NCCs and cardiomyocytes (CM). Consequently, loss of HMG20A, which marks open promoters and enhancers, results in chromatin accessibility changes and a striking deregulation of transcription programs involved in epithelial-mesenchymal transition (EMT) and differentiation processes. Collectively, our findings implicate HMG20A as part of the H2A.Z/PWWP2A/NuRD-axis and reveal it as a key modulator of intricate developmental transcription programs that guide the differentiation of NCCs and CMs.
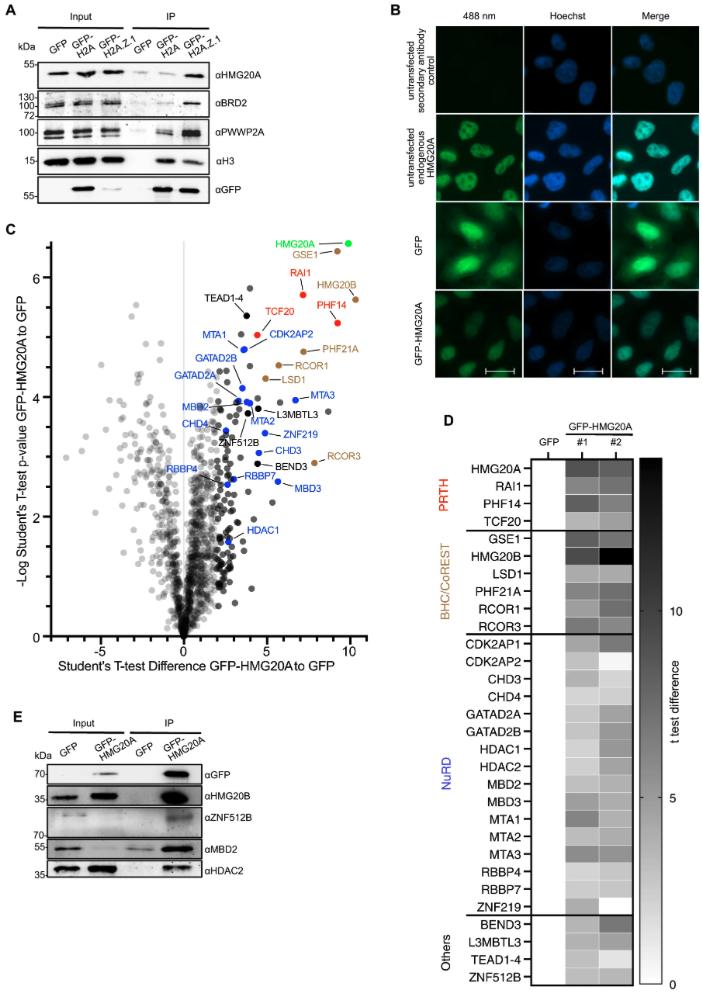
Fig. 1: HMG20A binds several chromatin-modifying complexes. A Immunoblots of GFP, HMG20A, BRD2 and PWWP2A (positive controls) as well as H3 upon GFP, GFP-H2A and GFP-H2A.Z.1 mononucleosome IPs. B Immunofluorescence microscopy images of GFP, GFP-HMG20A and endogenous HMG20A proteins (488 nm, green) in HeLaK cells. DNA is stained with Hoechst (blue). Scale bar: 20 µm. C Volcano plot of label-free interaction partners of GFP-HMG20A-associated mononucleosomes. Significantly enriched proteins over GFP-associated mononucleosomes are shown in the upper right part. t-Test (two-tailed) differences were obtained by two-sample t-test. HMG20A is highlighted in bright green, PRTH members in red, BHC/CoREST members in brown, NuRD members in blue, other proteins in black and background binding proteins in grey. See also Supplementary Fig. 1E for Volcano plot of second biological replicate and Supplementary Data 1 for detailed list of HMG20A binders. D Heatmap of significant outliers from two independent GFP-HMG20A mononucleosome IPs analysed by lf-qMS/MS (see C and Supplementary Fig. 1E) normalized to GFP. Scale bar: log2-fold t-test differences (two-tailed). E Immunoblots of GFP and GFP-HMG20A mononucleosome-IPs detecting endogenous members of the BHC/CoREST (HMG20B) and NuRD (MBD2, HDAC2) complexes as well as ZNF512B protein. Experiments in A, B, E were repeated independently three times with consistency. Source data for these figures are provided as a Source Data file.
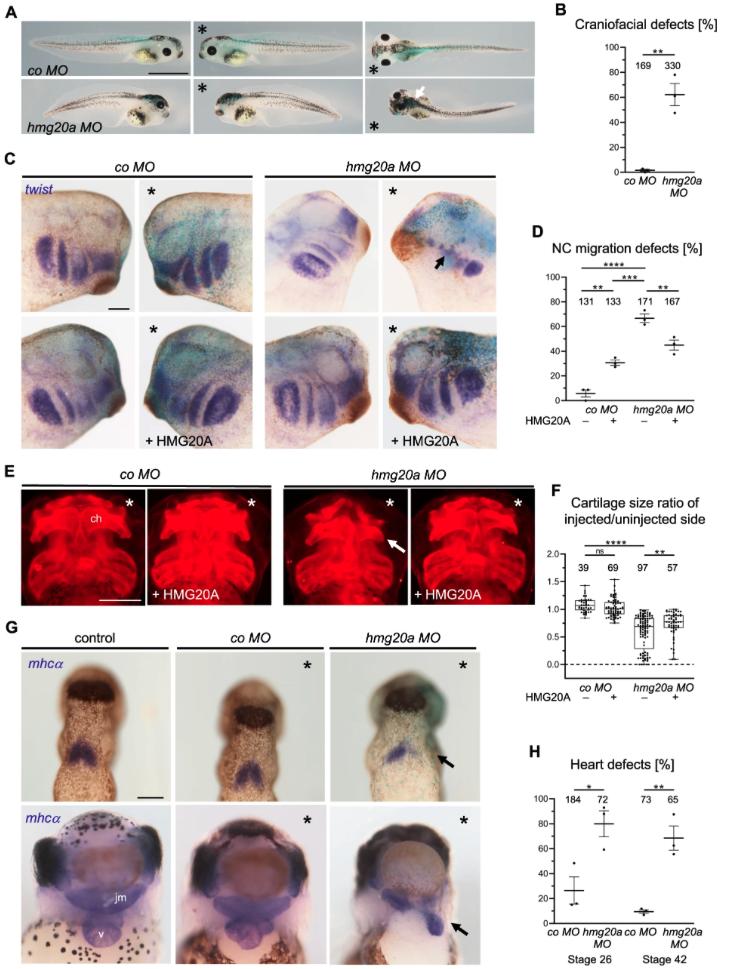
Fig. 4: HMG20A depletion leads to craniofacial and heart malformations in frog. A Loss of function of Hmg20a leads to craniofacial and pigmentation defects in Xenopus tadpoles. *Marks the injected side, white arrow marks pigmentation defects. Scale bar = 1 mm. B Mean percentage of craniofacial defects of three independent experiments ± s.e.m. Number of embryos are indicated for each column. **p = 0.0024 (two-tailed unpaired Student’s t-test). C Hmg20 loss-of-function NC migration defects can partially be rescued by co-injection of human HMG20A DNA. * Marks the injected side (blue: lacZ staining, purple: twist staining), arrow indicates cranial NC migration defect. Scale bar = 1 mm. D NC migration defects of three independent experiments injected and analysed as shown in (C). Data are presented as mean ± s.e.m., **p (left) = 0.0026, ***p = 0.0002 ****p = 0.0001, **p (right) = 0.006 (one-way ANOVA, Tukey’s multiple comparisons test). E Hmg20a-depleted Xenopus tadpole embryos show defects in cartilage formation (arrow). For rescue experiments, human HMG20A DNA was co-injected, *marks the injected side. Scale bar = 500 µm. F Box and whiskers plots summarize cartilage defects of at least three independent experiments analysed as in (E). Number of embryos (n, above each bar) and median are indicated. The box extends from 25th to 75th percentile, with whiskers from minimum to maximum. **p = 0.0013, ****p = 0.0001, ns.: not significant (one-way ANOVA, Tukey’s multiple comparisons test). G Hmg20a loss-of-function causes heart defects. Top: mhcα in situ hybridization reveals defects in the formation of the first heart field (arrow) at stage 26. Bottom: At stage 42, the three-chambered heart structure consisting of two atria (a) and a ventricle (v) is disturbed; the malformed heart is displaced towards the injected side (arrow). The jaw muscle (jm), which is also marked by mhcα, is also reduced. Scale bar = 1 mm. H Graph summarizing three independent experiments as shown in (G), data are presented as mean ± s.e.m. *p = 0.0242, **p = 0.0038 (two-tailed unpaired Student’s t-test). Source data for these figures are provided as a Source Data file.
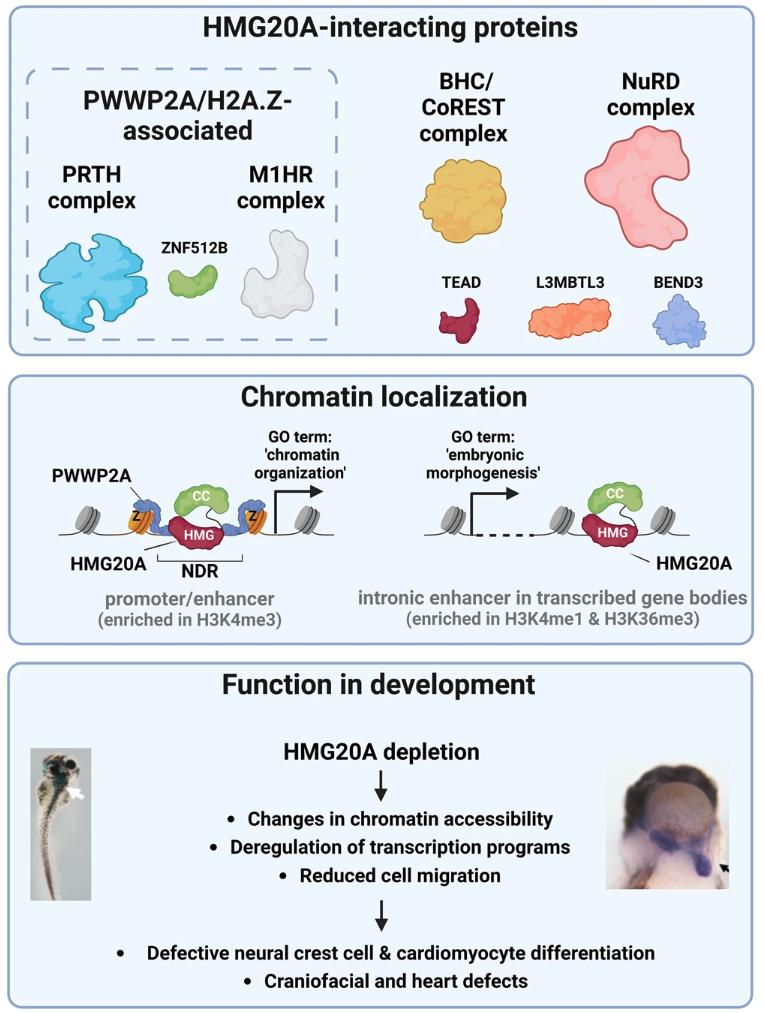
Fig. 9: Model of HMG20A’s function in chromatin and transcriptional regulation during development. Top: HMG20A associates with H2A.Z- and PWWP2A-associated PRTH and M1HR complexes and ZNF512B as well as BHC/CoREST and NuRD complexes and TEAD and L3MBTL3 that are not part of H2A.Z or PWWP2A interactomes. Middle: HMG20A binds to two distinct chromatin regulatory elements: (1) Nucleosome depleted regions (NDR) at promoter sites that are surrounded by H2A.Z-containing nucleosomes and bound by PWWP2A and that are associated with genes involved in basic processes, such as ‘chromatin organization‘. (2) H2A.Z-lacking intronic enhancers within transcribed genes belonging to developmental processes, such as ‘embryonic morphology”. Bottom: Depletion of HMG20A in Xenopus laevis and mESCs leads to changes in chromatin accessibility, deregulation of transcription programs as well as migration defects. HMG20A depleted cells fail to properly differentiate into neural crest cells or cardiomyocytes in mESCs as well as head and heart in Xenopus laevis. Figure was created with BioRender.
Adapted with permission from Springer Nature on behalf of Nature Communications: Herchenröther et al. (2023). The H2A.Z and NuRD associated protein HMG20A controls early head and heart developmental transcription programs. Nat Commun. 2023 Jan 28;14(1):472. doi: 10.1038/s41467-023-36114-x.
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
Last Updated: 2023-04-11