Cell landscape of larval and adult Xenopus laevis at single-cell resolution
Yuan Liao, Lifeng Ma, Qile Guo, Weigao E, Xing Fang, Lei Yang, Fanwei Ruan, Jingjing Wang, Peijing Zhang, Zhongyi Sun, Haide Chen, Zhongliang Lin, Xueyi Wang, Xinru Wang, Huiyu Sun, Xiunan Fang, Yincong Zhou, Ming Chen, Wanhua Shen, Guoji Guo & Xiaoping Han
Nat Commun. 2022 Jul 25;13(1):4306. doi: 10.1038/s41467-022-31949-2.
Click here to view article at Nature Communications.
Click here to view article on PubMed.
Click here to view article on Xenbase.
Click here to go to the Xenopus Cell Landscape resource.
Abstract
The rapid development of high-throughput single-cell RNA sequencing technology offers a good opportunity to dissect cell heterogeneity of animals. A large number of organism-wide single-cell atlases have been constructed for vertebrates such as Homo sapiens, Macaca fascicularis, Mus musculus and Danio rerio. However, an intermediate taxon that links mammals to vertebrates of more ancient origin is still lacking. Here, we construct the first Xenopus cell landscape to date, including larval and adult organs. Common cell lineage-specific transcription factors have been identified in vertebrates, including fish, amphibians and mammals. The comparison of larval and adult erythrocytes identifies stage-specific hemoglobin subtypes, as well as a common type of cluster containing both larval and adult hemoglobin, mainly at NF59. In addition, cell lineages originating from all three layers exhibits both antigen processing and presentation during metamorphosis, indicating a common regulatory mechanism during metamorphosis. Overall, our study provides a large-scale resource for research on Xenopus metamorphosis and adult organs.
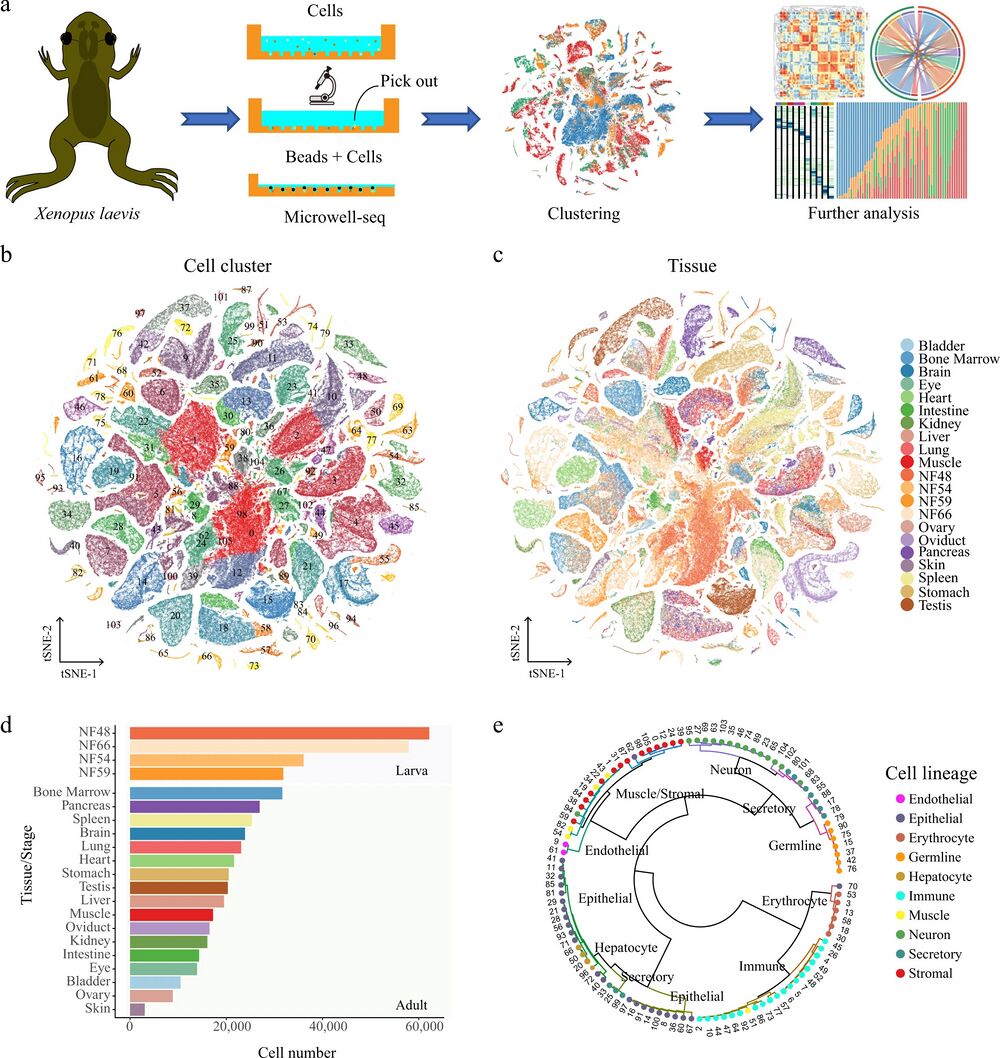
Fig. 1: Constructing an XCL using Microwell-seq.
a A schematic of the basic workflow for the Xenopus cell landscape using the Microwell-seq platform. b t-SNE analysis of 501, 358 single cells collected from larval and adult tissues. The 106 cell-type clusters are labeled in different colors. Cell cluster markers are listed in Supplementary Dataset 1. c t-SNE analysis of 501, 358 single cells collected from larval and adult tissues. Tissues/stages are labeled in different colors. d Cell number of each tissue/stage is detected at the XCL. Tissues/stages are labeled in different colors. e The 106 cell-type clusters are re-clustered into ten cell lineages and circles showing the relationships among the 106 cell types. Cell lineages are labeled in different colors.
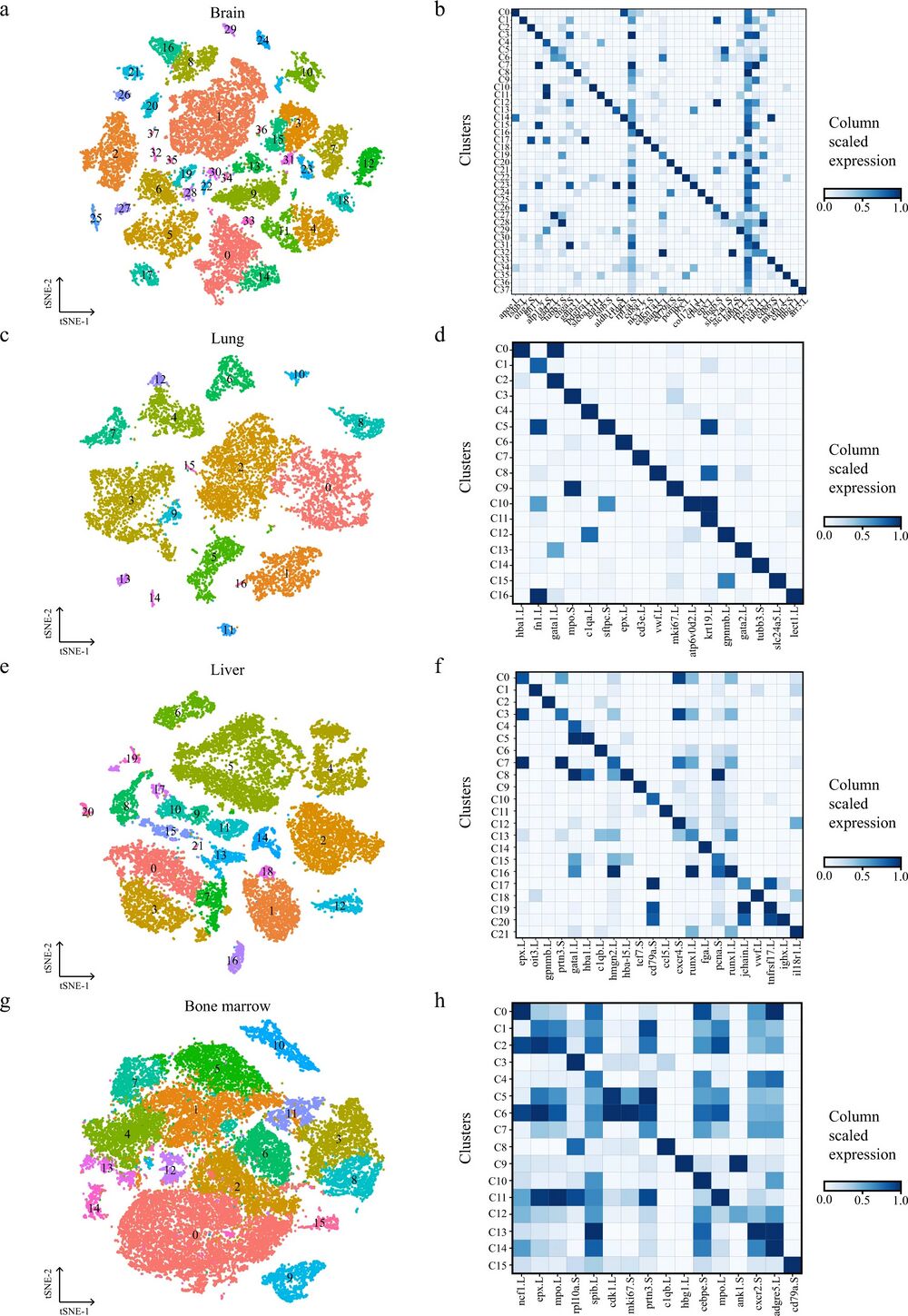
Fig. 2: Cellular heterogeneity in diverse adult Xenopus laevis tissues.
t-SNE map of Xenopus brain (a), lung (c), liver (e), and bone marrow (g) single-cell data. Cells are colored by cell-type cluster; Heatmap showing representative gene expression in each cluster of Xenopus brain (b), lung (d), liver (f), and bone marrow (h). The color encodes the average expression level.
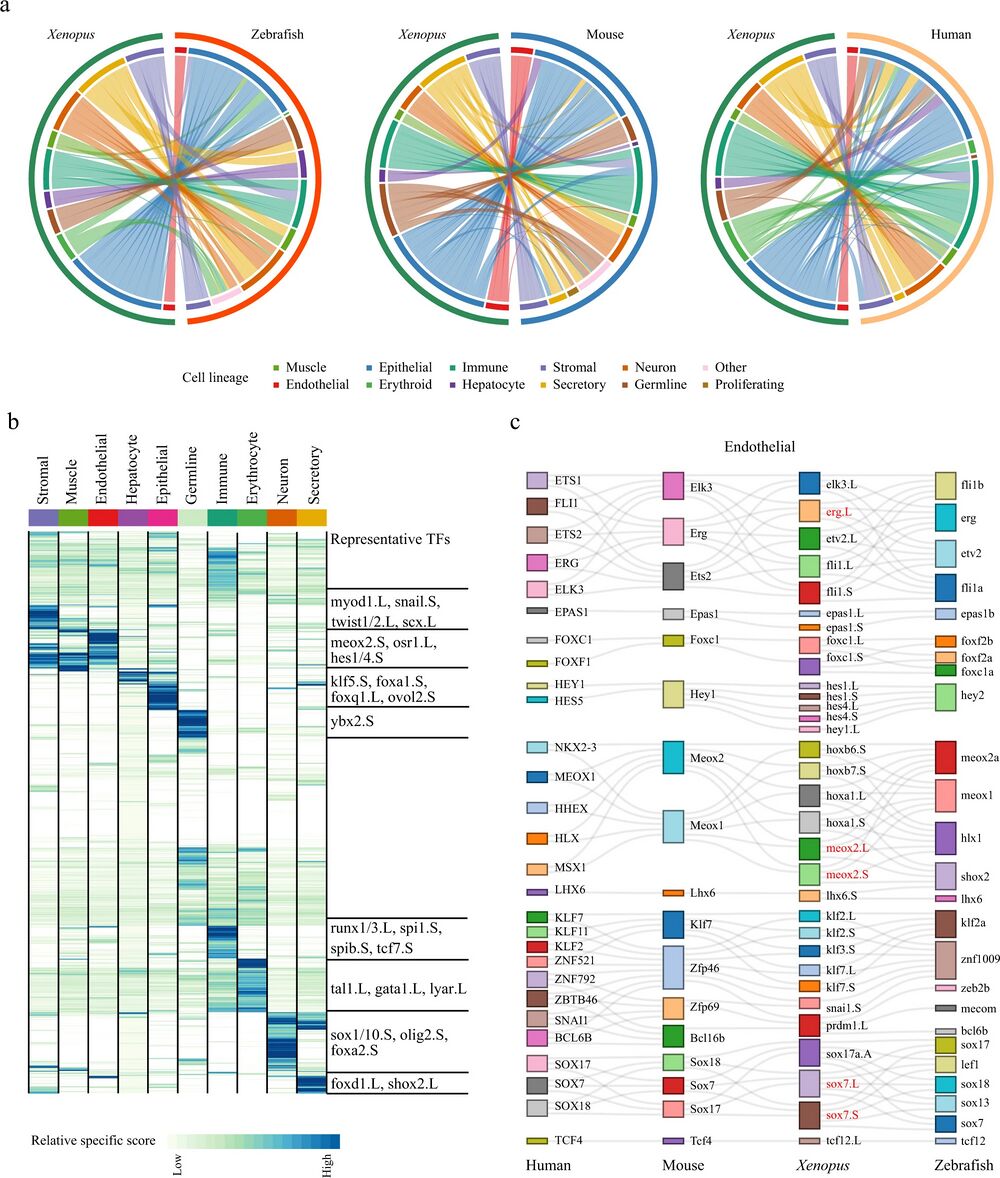
Fig. 3: Cell-type evolution between adult Xenopus, zebrafish, and mammals.
a Circle plot showing the similarity of cell lineages in Xenopus laevis, zebrafish (left), mice (middle), and humans (right). Pairs of cell types with mapping scores greater than 0.1 are connected by lines. Cell lineages are labeled in different colors. b Heatmap showing the specific scores of TFs in Xenopus for each cell type. Each row represents one TF, and each column represents one cell lineage. Cell lineages are labeled in different colors. The representative TFs in each cell lineage are presented in the right panel. c Sankey plot showing common lineage-specific transcription factors in endothelial for four vertebrates (humans, mice, Xenopus, and zebrafish). The representative TFs in Xenopus laevis are labeled by red. Different TFs in each species are marked in different color boxes. Homologous TFs between species obtained from SAMap are connected by lines.
Adapted with permission from Springer Nature on behalf of Nature Communications: Liao et al. (2022). Cell landscape of larval and adult Xenopus laevis at single-cell resolution. Nat Commun. 2022 Jul 25;13(1):4306. doi: 10.1038/s41467-022-31949-2.
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
Last Updated: 2022-09-21