Xenopus stem cells used to build living programmable organisms
A scalable pipeline for designing reconfigurable organisms
Sam Kriegman, Douglas Blackiston, Michael Levin, and Josh Bongard
PNAS published January 13, 2020. https://doi.org/10.1073/pnas.1910837117
Click here to view article at PNAS.
Click here to view article on PubMed.
Significance
Most technologies are made from steel, concrete, chemicals, and plastics, which degrade over time and can produce harmful ecological and health side effects. It would thus be useful to build technologies using self-renewing and biocompatible materials, of which the ideal candidates are living systems themselves. Thus, we here present a method that designs completely biological machines from the ground up: computers automatically design new machines in simulation, and the best designs are then built by combining together different biological tissues. This suggests others may use this approach to design a variety of living machines to safely deliver drugs inside the human body, help with environmental remediation, or further broaden our understanding of the diverse forms and functions life may adopt.
Watch movies detailing this study:
Movie 1: https://movie-usa.glencoesoftware.com/video/10.1073/pnas.1910837117/video-1
Movie 2: https://movie-usa.glencoesoftware.com/video/10.1073/pnas.1910837117/video-2
Abstract
Living systems are more robust, diverse, complex, and supportive of human life than any technology yet created. However, our ability to create novel lifeforms is currently limited to varying existing organisms or bioengineering organoids in vitro. Here we show a scalable pipeline for creating functional novel lifeforms: AI methods automatically design diverse candidate lifeforms in silico to perform some desired function, and transferable designs are then created using a cell-based construction toolkit to realize living systems with the predicted behaviors. Although some steps in this pipeline still require manual intervention, complete automation in future would pave the way to designing and deploying unique, bespoke living systems for a wide range of functions.
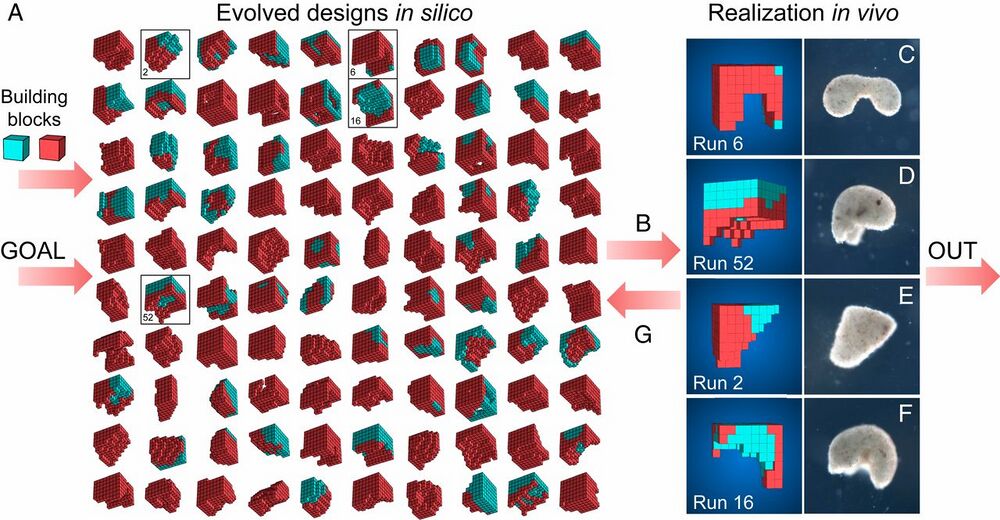
Fig. 1.
Designing and manufacturing reconfigurable organisms. A behavioral goal (e.g., maximize displacement), along with structural building blocks [here, contractile (red) and passive (cyan) voxels], are supplied to an evolutionary algorithm. The algorithm evolves an initially random population and returns the best design that was found. The algorithm is rerun 99 times starting with different random populations, generating a diversity of performant designs in silico (A; SI Appendix, section S5). Performant designs are then filtered by their robustness to random phase modulation of their contractile cells (B; SI Appendix, section S7), constructed in vivo using developing Xenopus cardiomyocyte and epidermal cell progenitors (C–F; SI Appendix, section S8), and placed on the surface of a Petri dish where their behavior is observed and compared to the design’s predicted behavior (SI Appendix, section S9). Discrepancies between in silico and in vivo behavior are returned to the evolutionary algorithm in the form of constraints on the kinds of designs that can evolve during subsequent design–manufacture cycles (G; SI Appendix, section S6). Concurrently, tissue layering and shaping techniques are modified such that realized living systems behave more like their virtual model (SI Appendix, section S8).
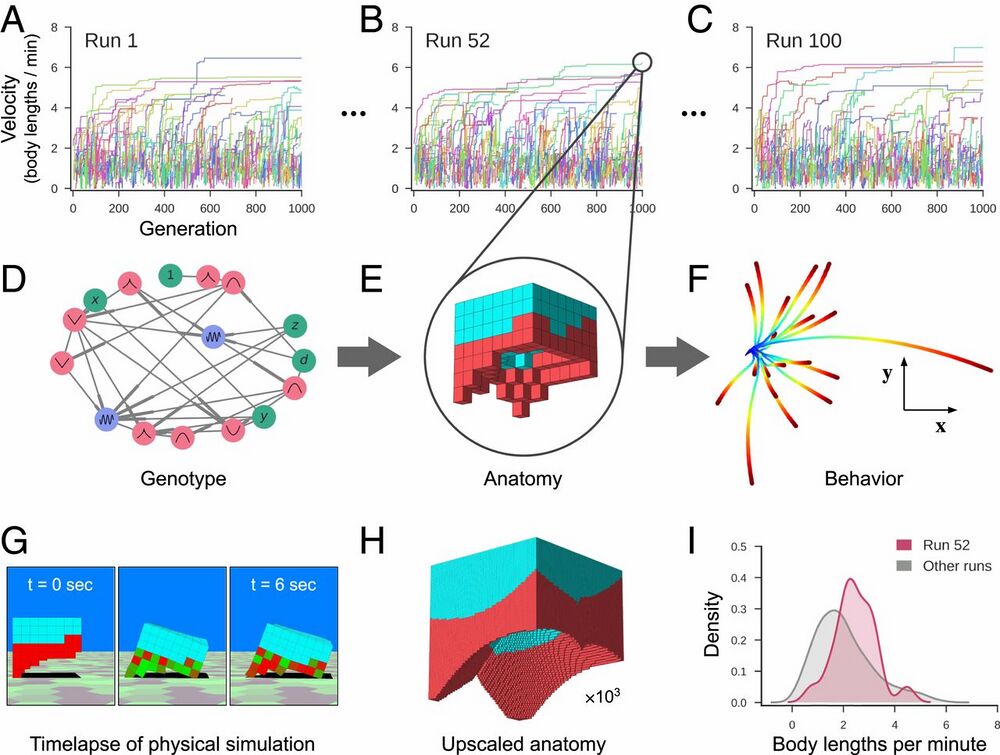
Fig. 2.
Designing reconfigurable organisms. For a given goal, 100 independent evolutionary trials were conducted in silico (A–C). Each colored line represents the velocity of the fastest-moving design within its clade. Each genome (D) dictates anatomy and behavior by determining where and how voxels are combined, and whether they are passive (cyan) or contractile (red; E). Genomes simulate a developmental process and are described in more detail in SI Appendix, section S4. The differing behavioral traces produced by a design (F) are a result of randomly perturbing the actuation of each contractile cell during each evaluation period. The behavioral traces all originate from the same position (blue) but diverge over time until their final destination (red). (G) During one evaluation period, after settling under gravity for 1 s, compressed and expanded contractile voxels are shown in red and green, respectively. Because the genotype is scale-free, the anatomical resolution of any design can be increased (H) while preserving geometry (but not necessarily behavior). When all evolutionary trials complete, the most performant design from each trial is extracted (I). The robust design passed to the next stage of the pipeline moves, on average, more rapidly (red curve) than the average speed of the other 99 designs (gray curve).
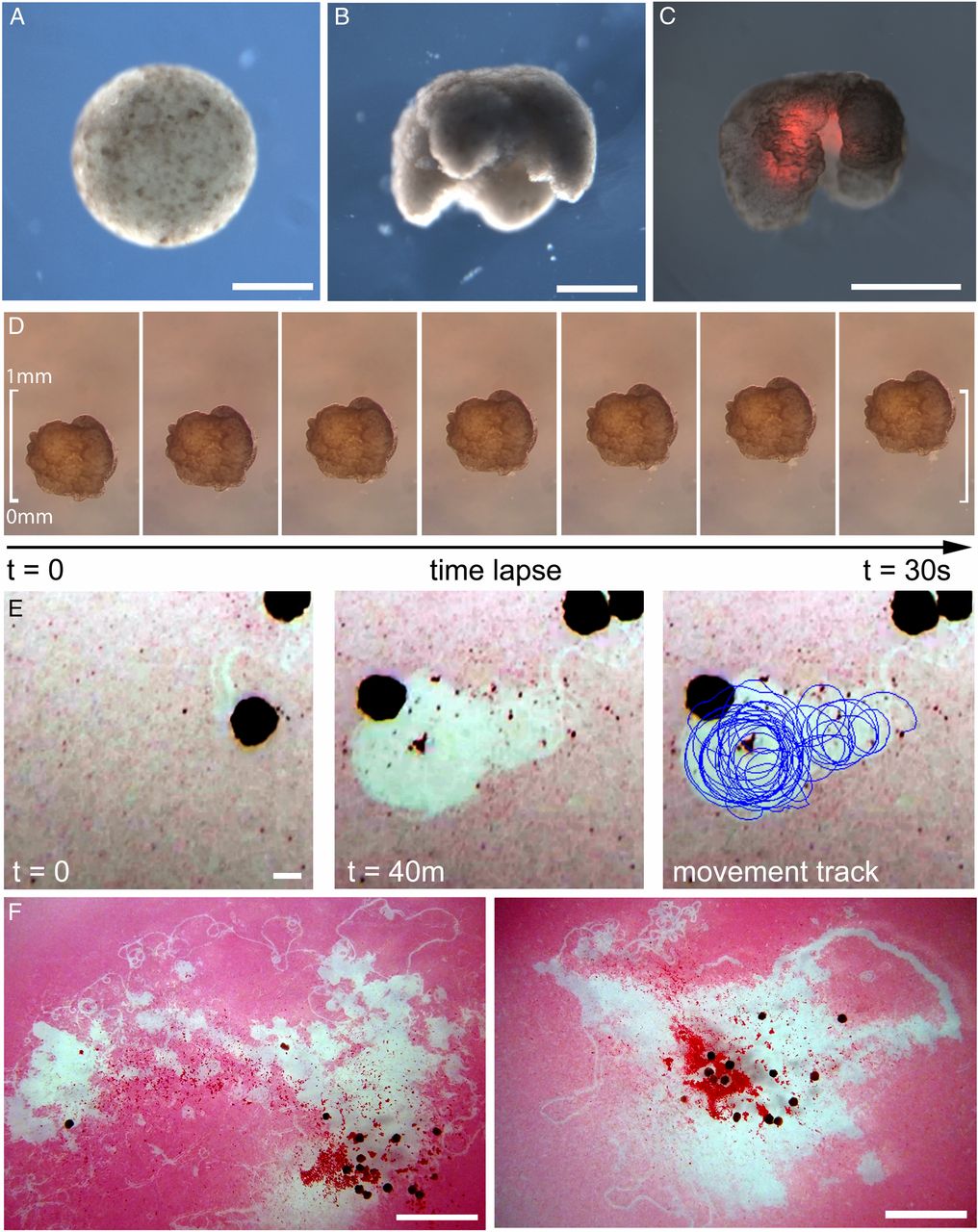
Fig. 3.
Manufacturing reconfigurable organisms. (A) Aggregation of pluripotent blastula cells harvested from X. laevis embryos. (B) Shaping results in 3D representations of the evolved in silico designs. (C) Layering of cardiac progenitor cells results in contractile cardiomyocyte tissue at specific locations, visualized by red fluorescent lineage tracer. (D) Time-lapse imaging of self-locomotion in an aqueous environment. (E) Emergent behavior of debris aggregation by an individual within the environment and (F) by groups of reconfigurable organisms over a 24-h period (SI Appendix, section S10.4). (Scale bars: 500 μm for A–E and 5 mm for F, respectively.)
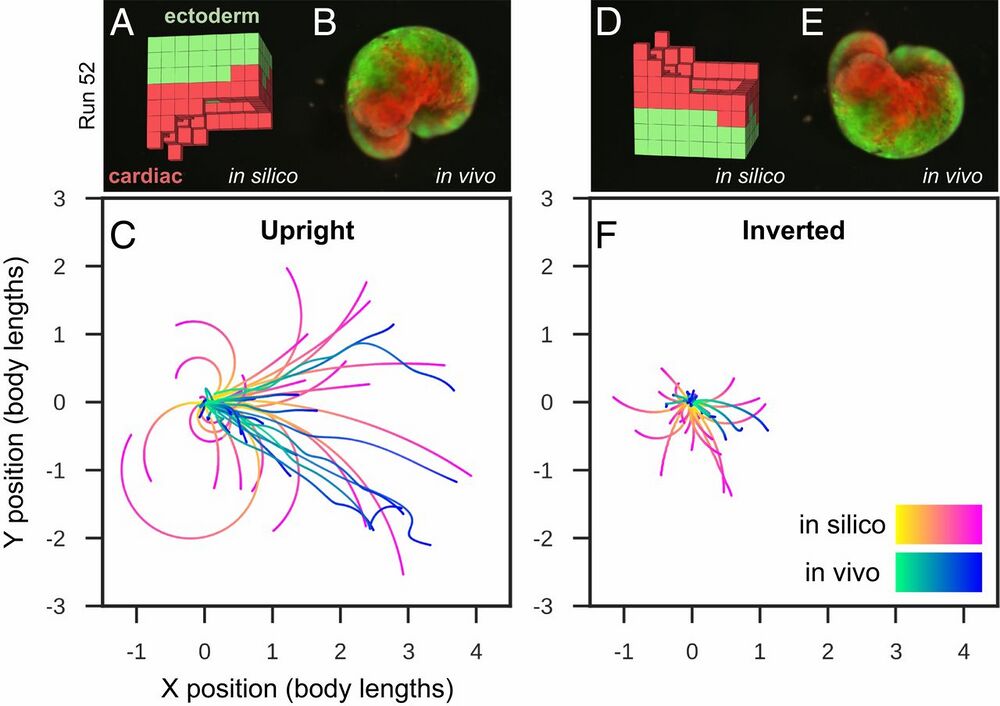
Fig. 4.
Transferal from silico to vivo. The first design selected for fabrication and specific hypothesis testing (A) was the most robust yet stable and energy-efficient configuration of passive (epidermis; green) and contractile (cardiac; red) tissues found by the evolutionary algorithm. The design was evaluated 25 times for 1 min of simulation time, resulting in 25 movement trajectories (pink curves in C). Six reconfigurable organisms were built which embodied this design (e.g., B) (SI Appendix, section S9). Three were evaluated four times and the other three were evaluated five times for 10 min each (27 blue curves in C). The organisms’ direction of movement matched the design’s predicted direction of movement (P < 0.01; details in SI Appendix, section S9). To determine whether the organisms’ movement was a result of chance or due to the design’s evolved geometry and tissue placement, geometry and tissue distribution was altered by rotating the design 180° about its transverse plane (D) and evaluating it another 25 times in silico (pink curves in F). Each of the six organisms were likewise inverted (E): four were evaluated five times while the remaining two were only evaluated once (22 blue curves in F). Inverting the design significantly reduces its net displacement (P < 0.001), as did inverting the organisms (P < 0.0001).
Adapted with permission from the National Academy of Sciences: Kriegman et al. (2020). A scalable pipeline for designing reconfigurable organisms.
PNAS first published January 13, 2020. https://doi.org/10.1073/pnas.1910837117 Copyright (2020).
