Cell & Developmental Biology of Xenopus: Gene Discovery & Disease 2020 Course at Cold Spring Harbor
https://meetings.cshl.edu/xeno20

Click here for the pdf of the poster.
Cell and Developmental Biology of Xenopus: 2019 Course at Cold Spring Harbor
Cell & Developmental Biology of Xenopus:
Gene Discovery & Disease
March 25 - April 7, 2020
Application & Materials Deadline: February 11, 2020
Instructors:
Lance Davidson, University of Pittsburgh
Chenbei Chang, University of Alabama at Birmingham
See the roll of honor - who's taken the course in the past.
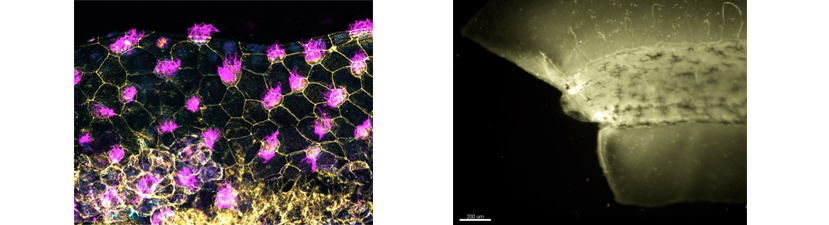
In vivo animal models are an important tool for the understanding of human development and disease. Studies using the frog Xenopus have made remarkable contributions to our understanding of fundamental processes such as cell cycle regulation, transcription, translation and many other topics. Xenopus is remarkable for studying development and disease, including birth defects, cancer, and stem cell biology. Because Xenopus are easy to raise, producing many thousands of eggs per day, these frogs have emerged as a premiere model for understanding of human biology from the fundamental building blocks to the whole organism.
The recent development of CRISPR/Cas9 technology has made it easy to target genes of interest using Xenopus. This course has been designed with that in mind. Our goal is for each student to design a set of experiments focusing on their gene or biological interest. Prior to starting the course, students will be expected to choose gene(s) of interest, and the instructors will generate sgRNAs targeting these genes. These can be the students’ own genes, or chosen from a bank provided by the instructors. The gene targeting experiments will be combined with other manipulations, such as tissue explants and transplants and live imaging to analyze the function of the genes.
Xenopus is increasingly being used as imaging test-bed to investigate the roles of cytoskeleton and intracellular trafficking in cell biological and morphogenetic contexts. The course maintains stock mRNAs for targeting fluorescent proteins to specific structures for studying cell shape and cytoskeletal dynamics but students are encouraged to bring or suggest additional tools, including fluorescent biosensors, tension-sensors, etc. The power of Xenopus can be leveraged when live-cell fluorescence imaging is combined with microsurgery, grafting, and dissociated cell culture.
During the course, the students will analyze any phenotypes generated from CRISPR/Cas9 based gene depletion while learning the diverse array of techniques available in Xenopus. In previous courses, we have guided students in the ablation of a wide variety of genes and helped them design suitable assays for their biological interests. Most recently, students have targeted autism genes, thyroid genes and immune modulators, several of which have already led to publications. Approaches covered will include microinjection and molecular manipulations such as CRISPR/Cas9 knockouts, antisense morpholino-based depletions, transgenics, and mRNA overexpression. In addition, students can combine these techniques with explant and transplant methods to simplify or test tissue level interactions. Additional methods include mRNA in situ hybridization and protein immunohistochemistry as well as basic bioinformatic techniques for gene comparison and functional analysis. Biochemical approaches such as proteomics and mass spectrometry and biomechanical concepts will also be discussed. Finally, to visualize subcellular and intercellular activities, we will introduce a variety of sample preparation and imaging methods including time-lapse, fluorescent imaging, optical coherence tomography and confocal microscopy. These are facilitated by state-of-the-art equipment from Nikon, Leica, Thorlabs, and Bruker.
Due to the tailored nature of this course, it is suitable for those new to the Xenopus field, as well as for more advanced students who are interested in emerging technologies. Please feel free to contact the instructors for informal guidance.
2020 Lecturers:
Rebecca Heald, University of California, Berkeley
Douglas Houston, University of Iowa
Ray Keller, University of Virginia
Mustafa Khokha, Yale University
Roberto Mayor, University College London, UK
Brian Mitchell, Northwestern University
Nanette Nascone-Yoder, North Carolina State University
Andrea Wills, University of Washington
Sarah Woolner, University of Manchester, UK
Martin Wuhr, Princeton University
Please visit the course website to apply.
Last Updated: 2020-02-04