Defining the vertebrate cardiac proteome
This work identified and characterized the core protein composition, protein complexes and protein pathways in vertebrates from Xenopus to human. Surprisingly, the authors also revealed that each species shares a unique set of proteins with humans but not the other vertebrates studied, and that these proteins are mutated in human disease states. The studies therefore, emphasize the use of specific model systems for a defined biological process and allow a prediction a priori of the appropriate animal model for studying specific human cardiac conditions.
Conservation and divergence of protein pathways in the vertebrate heart
Joel D. Federspiel, Panna Tandon, Caralynn M. Wilczewski, Lauren Wasson, Laura E. Herring, Samvida S. Venkatesh, Ileana M. Cristea, Frank L. Conlon
PLoS Biol. 2019 Sep 6;17(9):e3000437. doi: 10.1371/journal.pbio.3000437.
Click here to view article at PLoS Biology (figures, supplemental figures, tables, and movies).
Click here to view article on Pubmed.
Click here to view article on Xenbase (figures and supplemental figures).
Abstract
Heart disease is the leading cause of death in the western world. Attaining a mechanistic understanding of human heart development and homeostasis and the molecular basis of associated disease states relies on the use of animal models. Here, we present the cardiac proteomes of 4 model vertebrates with dual circulatory systems: the pig (Sus scrofa), the mouse (Mus musculus), and 2 frogs (Xenopus laevis and Xenopus tropicalis). Determination of which proteins and protein pathways are conserved and which have diverged within these species will aid in our ability to choose the appropriate models for determining protein function and to model human disease. We uncover mammalian- and amphibian-specific, as well as species-specific, enriched proteins and protein pathways. Among these, we find and validate an enrichment in cell-cycle-associated proteins within Xenopus laevis. To further investigate functional units within cardiac proteomes, we develop a computational approach to profile the abundance of protein complexes across species. Finally, we demonstrate the utility of these data sets for predicting appropriate model systems for studying given cardiac conditions by testing the role of Kielin/chordin-like protein (Kcp), a protein found as enriched in frog hearts compared to mammals. We establish that germ-line mutations in Kcp in Xenopus lead to valve defects and, ultimately, cardiac failure and death. Thus, integrating these findings with data on proteins responsible for cardiac disease should lead to the development of refined, species-specific models for protein function and disease states.
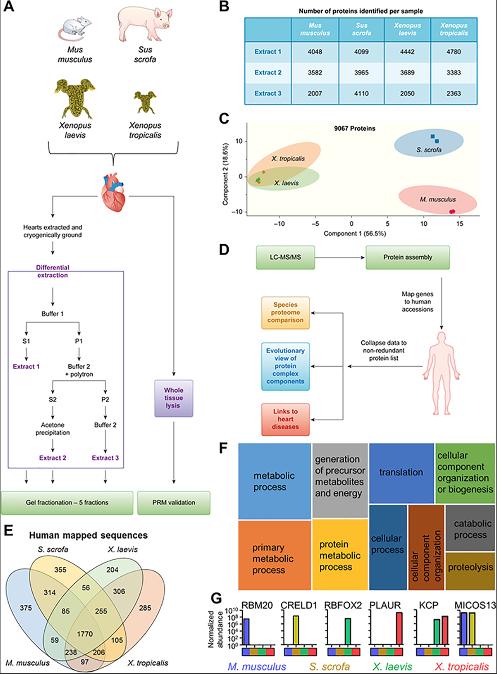
Fig 1. Workflow for investigating multispecies cardiac proteomes. (A) Heart tissue from M. musculus, S. scrofa, X. laevis, and X. tropicalis was collected and subjected to differential protein extraction as detailed. The resulting 3 extracts were fractionated by SDS-PAGE. (B) Number of proteins identified in the 5 gel fractions for each extract (extract 1–3 from panel A) per species. These numbers include proteins found in 2 or more extracts. (C) PCA of MS1 proteome data demonstrates that the mammalian samples separate from each other and the Xenopus samples but the Xenopus samples maintain a tight association. (D) Data analysis workflow. (E) Identified proteins were mapped to human accession numbers using Blast2GO. (F) The shared proteins in all species represent a core cardiac proteome that is enriched for a variety of pathways. Shown here in a tree map are the top 10 most enriched (adjusted p ≤ 0.05) GO biological process terms; box size scales with enrichment significance of the terms. (G) Examples of proteins detected in subsets of the species analyzed. See S8 Table for numerical data underlying figure. GO, Gene Ontology; Kcp, Kielin/chordin-like protein; LC-MS/MS, liquid chromatography coupled with tandem mass spectrometry;MS1, precursor ion.
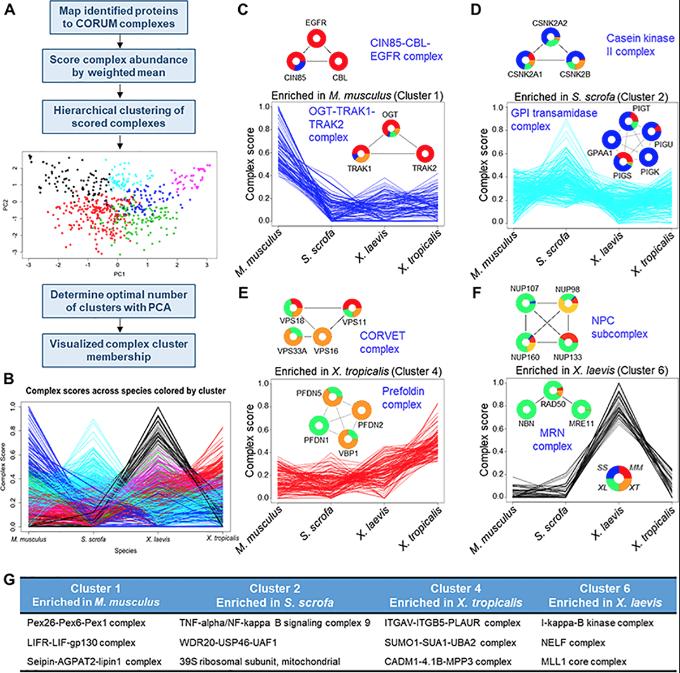
Fig 2. Evolutionary comparison of protein complexes. (A) Proteins detected in this study were mapped to known human protein complexes listed in CORUM. (B) A complex score was calculated from the MS1 peak area of individual protein components of each complex. These complexes were then clustered into 6 clusters and analyzed further. (C–F) Individual clusters are plotted along with example protein complexes, demonstrating the underlying abundance data that drove the complex clustering. (G) Three representative complexes for each cluster are listed. See S9 Table for numerical data underlying figure. CORUM, Comprehensive Resource of Mammalian Protein Complexes.
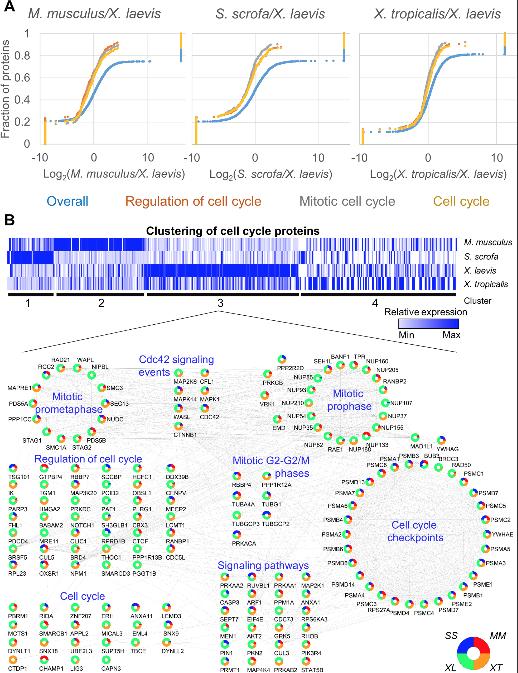
Fig 3. GSEA reveals the increased relative abundance in cell-cycle proteins in Xenopus laevis. (A) Pairwise GSEA was carried out between X. laevis and the 3 other species, revealing an enrichment (adjusted p ≤ 0.05) in cell cycle related proteins in X. laevis. (B) All proteins found in any enriched cell cycle related category by GSEA were clustered using k means (k = 4). Relative protein abundance is shown. The proteins found in cluster 3 were further analyzed in Cytoscape and grouped by function to show the interrelated nature of the proteins enriched in this cluster. See S13 Table and S14 Table for numerical data underlying figure. GSEA, Gene Set Enrichment Analyses.
Adapted with permission from PLOS Biology Federspiel et al. (2019). Conservation and divergence of protein pathways in the vertebrate heart. PLOS Biol 2019 Sept 6; 17(9):e3000437. doi: 10.1371/journal.pbio.3000437. Copyright (2019).
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
Last Updated: 2019-09-11