Epigenome reference map track hub
2/8/2016
Track hubs now include trop v9.1 assembly.
UCSC trackhub information: http://genome.ucsc.edu/goldenPath/help/hgTrackHubHelp.html
1/4/2015
Release of Xenopus tropicalis epigenome reference maps
A collection of over 70 epigenome reference maps of Xenopus tropicalis early development has been released with the publication of “Embryonic transcription is controlled by maternally defined chromatin state” in Nature Communications.
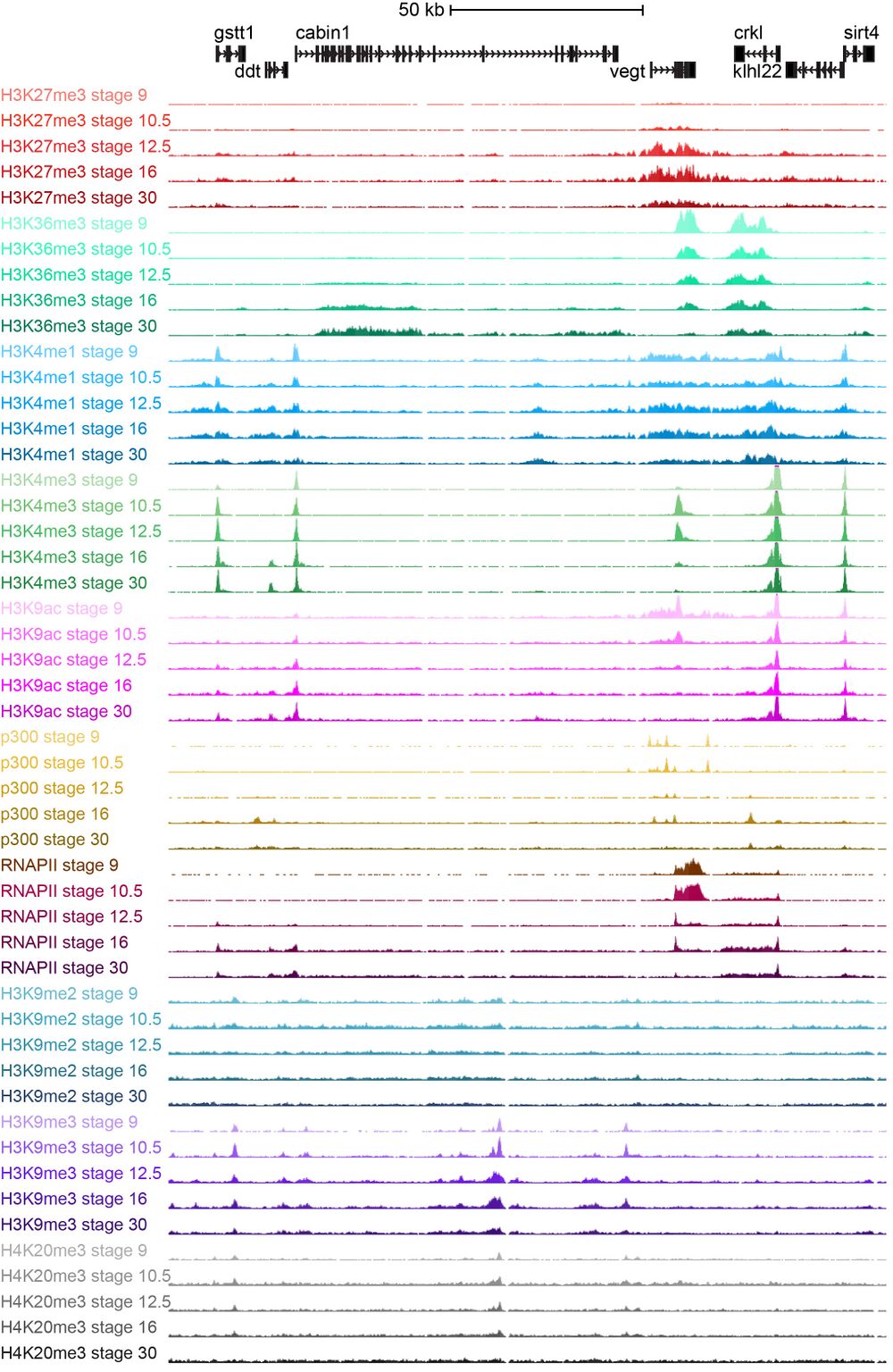
The data documents the genome-wide patterns of histone modifications (H3K4me1, H3K4me3, H3K9ac, H3K9me2, H3K9me3, H3K27me3, H3K36me3, H4K20me3), DNA methylation (WGBS-seq), and the occupancy of RNA polymerase II and the transcriptional co-activator p300 at developmental stages ranging from blastula to tailbud. The epigenetic marks reveal chromatin state dynamics at promoters and enhancers and show the patterns of facultative and constitutive heterochromatin. In addition the study has analyzed how embryonic transcription influences the acquisition of histone modifications using the RNA polymerase inhibitor alpha-amanitin. The results indicate a pervasive influence of maternal factors at the level of chromatin state, an influence that persists well into post-gastrulation development. The data also show that the maternal-to-zygotic transition is not so much a clean developmental transition in time, but rather involves a segregation of genomic space in maternal and zygotic spheres of influence.
The study was funded by NIH/NICHD grant 1R01HD069344. For the paper, access to data, genome browser tracks, and more information:
· Embryonic transcription is controlled by maternally defined chromatin state. 2015. Hontelez S, van Kruijsbergen I, Georgiou G, van Heeringen SJ, Bogdanovic O, Lister R, Veenstra GJ. Nat Commun. 2015 Dec 18;6:10148. doi:10.1038/ncomms10148. PubMed PMID: 26679111.
· A description of the track hub and step-by-step instructions on how to load the track hub can be found at the following location: http://veenstra.science.ru.nl/trackhubx.htm
· NCBI Geo: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE67974
~
3/2/2015
Work from the Veenstra lab funded under the NIH/NICHD grant, 1R01HD069344, shows over 50 genome browser tracks to be released for the Xenopus tropicalis v7.1 and v8.0 assemblies. These ChIP-sequencing tracks cover eight histone modifications, RNA polymerase II (RNAPII) and the enhancer-binding protein p300 during Xenopus tropicalis development from blastula to larval stages.
Description
The ChIP-sequencing tracks were produced as part of a project funded by NICHD to generate epigenome reference maps of Xenopus tropicalis embryos. Intergenic DNA contains functional genomic elements which are largely unannotated. The biochemical features of chromatin, such as post-translational modifications of histone proteins, determine its accessibility and function. The combination of different histone modifications can also be used to partition the genome in distinct chromatin states, such as promoters, enhancers, transcribed regions, and repressed heterochromatin.
ChIP-sequencing was performed as described (Akkers et al. 2009, Akkers et al. 2012, Van Heeringen et al. 2014). The genome browser tracks have been generated for the histone modifications H3K4me1, H3K4me3, H3K9ac, H3K9me2, H3K9me3, H3K27me3, H3K36me3 and H4K20me3, in addition to RNAPII and p300. At five stages of development (NF stages 9, 10.5, 12.5, 16 and 30) they have been mapped with or without repetitive reads, to allow assessing histone modfications at repetitive heterochromatic regions as well as unique regions within the genome. In addition they have been mapped to the two newest X.tropicalis genome assemblies (v7.1 and v8.0).
Using the track data hub
The data can be browsed using a new track data hub for the UCSC browser generated by the Veenstra lab. To use this track hub, visit the UCSC genome browser (http://genome.ucsc.edu/cgi-bin/hgGateway?org=X.+tropicalis) and add the track hub located at http://trackhub.science.ru.nl/hubs/xenopus/hub.txt
The hub contains several Xenopus genome assemblies as well as the new X. tropicalis data.
Credits and Data access policy
The data were generated in the Veenstra lab at Radboud University. The track hub includes published data but also provides pre-publication access to data. The unpublished data can be freely used on a per gene locus basis. Please contact Gert Jan Veenstra (email g.veenstra@science.ru.nl) for other use.
References
Akkers RC, Jacobi UG, Veenstra GJ. 2012. Chromatin immunoprecipitation analysis of Xenopus embryos. Methods Mol Biol. 2012;917:279-92. doi: 10.1007/978-1-61779-992-1_17. PMID: 22956095
Akkers RC, van Heeringen SJ, Jacobi UG, Janssen-Megens EM, Françoijs KJ, Stunnenberg HG, Veenstra GJ. 2009. A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev Cell. 2009 Sep;17(3):425-34. doi: 10.1016/j.devcel.2009.08.005. PMID: 19758566
van Heeringen SJ, Akkers RC, van Kruijsbergen I, Arif MA, Hanssen LL, Sharifi N, Veenstra GJ. 2014. Principles of nucleation of H3K27 methylation during embryonic development. Genome Res. 2014 Mar;24(3):401-10. doi: 10.1101/gr.159608.113. Epub 2013 Dec 11. PMID: 24336765
