CRISPr/Cas9-mediated targeted mutagenesis in Xenopus
Please follow this link for additional details on genome editing technologies.
Recently, targeted gene gene mutations using zinc-finger and transcription activator-like effector nucleases (ZFNs and TALENs) which function as dimers to bind targeted sites in the genome and cause site-specific double stranded breaks in the DNA. These breaks are repaired and mutations can be inserted at the site of repair.
Another simpler technology, CRISPr/Cas (clustered regularly interspaced short palindromic repeats-assiciated) is employed in these studies to induce targeted genetic mutation. This technology utilizes the Cas9 endonuclease from the bacterium Streptococcus pyogenes to the target site which normally utilizes two RNAs. CRISPr RNA (crRNA) has complementary sequence to the target gene DNA. A transactivating CRISPr RNA (tracrRNA) basepairs with crRNA. The target sequence must be followed by a tri-nucleotide protospacer adjacent motif (PAM) sequence - NGG. To perform targeted genetic mutation, these characteristic RNAs are combined synthetically in a single synthetic guide RNA (sgRNA). Co-injection of this RNA with Cas9 mRNA into early fertilized eggs functions to induces site-specific DNA strand breaks. DNA repair with added nucleotide bases complementary to the sgRNA sequence induces frameshift mutations.
In back-to-back articles published in genesis, Blitz et al. and Nakayama et al. each report CRISPr/Cas targeted transgenesis of Xenopus tropicalis pigmentation and eye development genes.
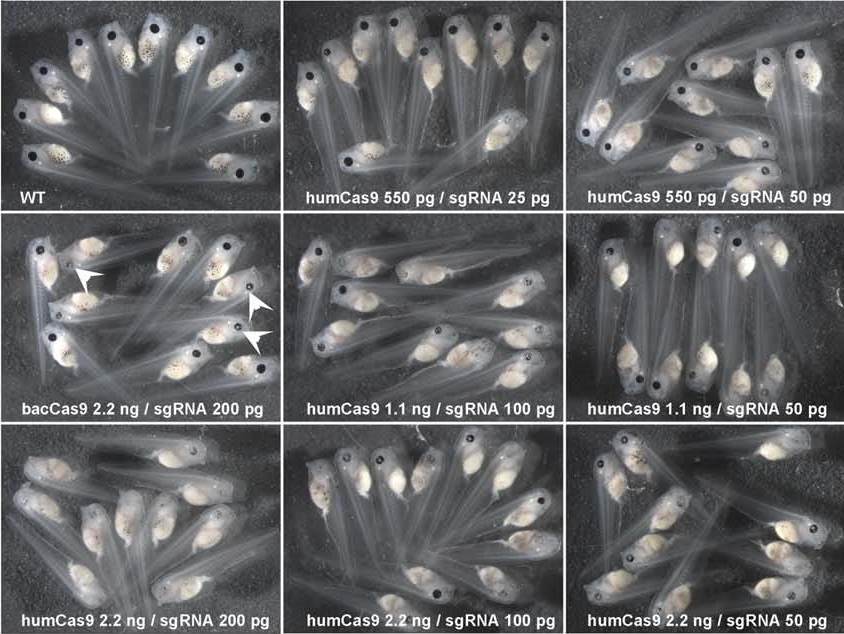
Biallelic genome modification in F0 Xenopus tropicalis embryos using the CRISPR/Cas system
The CRISPR/Cas system is adapted for use in Xenopus tropicalis and may be a useful system for creating genome modifications in this important model organism.
The Cho lab successfully targeted the tyrosinase (tyr) gene which confers pigmentation. Suppression of the tyrosinase gene results in albino frogs. A variable degree of mosaicism in the resulting animals. Albino F0 embryos are efficiently generated indicating that both tyr alleles are modified.
Published online November 8, 2013 in genesis. Blitz et al.
Simple and efficient CRISPR/Cas9-mediated targeted mutagenesis in Xenopus tropicalis
The Grainger lab successfuly targeted the tyrosinase (tyr) gene causing albinism in Xenopus embryos. The lack of pigmentation earlier in development is seen in the lack of pigment in the eye (retinal pigment epithelium) and in melanocytes. The eye development gene, six3 (an early eye field transcription factor) was also targeted for mutation and faulty eye development resulted in these mutants.
Published online November 8, 2013 in genesis. Nakayama et al.
Unpublished images provided by Ira Blitz and Takuya Nakayama.
Reviewed by VGP.
In back-to-back articles published in genesis, Blitz et al. and Nakayama et al. each report CRISPr/Cas targeted transgenesis of Xenopus tropicalis pigmentation and eye development genes.